Correcting errors in synthetic DNA through consensus shuffling
- PMID: 15800206
- PMCID: PMC1072806
- DOI: 10.1093/nar/gni053
Correcting errors in synthetic DNA through consensus shuffling
Abstract
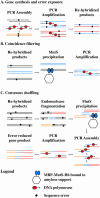
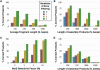
Although efficient methods exist to assemble synthetic oligonucleotides into genes and genomes, these suffer from the presence of 1-3 random errors/kb of DNA. Here, we introduce a new method termed consensus shuffling and demonstrate its use to significantly reduce random errors in synthetic DNA. In this method, errors are revealed as mismatches by re-hybridization of the population. The DNA is fragmented, and mismatched fragments are removed upon binding to an immobilized mismatch binding protein (MutS). PCR assembly of the remaining fragments yields a new population of full-length sequences enriched for the consensus sequence of the input population. We show that two iterations of consensus shuffling improved a population of synthetic green fluorescent protein (GFPuv) clones from approximately 60 to >90% fluorescent, and decreased errors 3.5- to 4.3-fold to final values of approximately 1 error per 3500 bp. In addition, two iterations of consensus shuffling corrected a population of GFPuv clones where all members were non-functional, to a population where 82% of clones were fluorescent. Consensus shuffling should facilitate the rapid and accurate synthesis of long DNA sequences.
Figures



References
-
- Matteucci M.D., Caruthers M.H. Nucleotide chemistry. 4. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1981;103:3185–3191.
-
- Singh-Gasson S., Green R.D., Yue Y.J., Nelson C., Blattner F., Sussman M.R., Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. - PubMed
-
- Stemmer W.P.C., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

