A computational study of off-target effects of RNA interference
- PMID: 15800213
- PMCID: PMC1072799
- DOI: 10.1093/nar/gki324
A computational study of off-target effects of RNA interference
Abstract
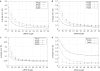
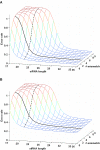
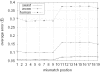
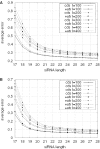
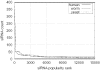
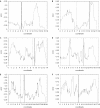
RNA interference (RNAi) is an intracellular mechanism for post-transcriptional gene silencing that is frequently used to study gene function. RNAi is initiated by short interfering RNA (siRNA) of approximately 21 nt in length, either generated from the double-stranded RNA (dsRNA) by using the enzyme Dicer or introduced experimentally. Following association with an RNAi silencing complex, siRNA targets mRNA transcripts that have sequence identity for destruction. A phenotype resulting from this knockdown of expression may inform about the function of the targeted gene. However, 'off-target effects' compromise the specificity of RNAi if sequence identity between siRNA and random mRNA transcripts causes RNAi to knockdown expression of non-targeted genes. The complete off-target effects must be investigated systematically on each gene in a genome by adjusting a group of parameters, which is too expensive to conduct experimentally and motivates a study in silico. This computational study examined the potential for off-target effects of RNAi, employing the genome and transcriptome sequence data of Homo sapiens, Caenorhabditis elegans and Schizosaccharomyces pombe. The chance for RNAi off-target effects proved considerable, ranging from 5 to 80% for each of the organisms, when using as parameter the exact identity between any possible siRNA sequences (arbitrary length ranging from 17 to 28 nt) derived from a dsRNA (range 100-400 nt) representing the coding sequences of target genes and all other siRNAs within the genome. Remarkably, high-sequence specificity and low probability for off-target reactivity were optimally balanced for siRNA of 21 nt, the length observed mostly in vivo. The chance for off-target RNAi increased (although not always significantly) with greater length of the initial dsRNA sequence, inclusion into the analysis of available untranslated region sequences and allowing for mismatches between siRNA and target sequences. siRNA sequences from within 100 nt of the 5' termini of coding sequences had low chances for off-target reactivity. This may be owing to coding constraints for signal peptide-encoding regions of genes relative to regions that encode for mature proteins. Off-target distribution varied along the chromosomes of C.elegans, apparently owing to the use of more unique sequences in gene-dense regions. Finally, biological and thermodynamical descriptors of effective siRNA reduced the number of potential siRNAs compared with those identified by sequence identity alone, but off-target RNAi remained likely, with an off-target error rate of approximately 10%. These results also suggest a direction for future in vivo studies that could both help in calibrating true off-target rates in living organisms and also in contributing evidence toward the debate of whether siRNA efficacy is correlated with, or independent of, the target molecule. In summary, off-target effects present a real but not prohibitive concern that should be considered for RNAi experiments.
Figures



References
-
- Fire A., Xu S.Q., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Fraser A.J.G., Kamath R.S., Zipperten P., Campos M.M., Sohrmann M., Ahringer J. Functional genomic analysis of C.elegans chromosome I by systemic RNA interference. Nature. 2000;408:325–330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

