A comparison of the binding sites of matrix metalloproteinases and tumor necrosis factor-alpha converting enzyme: implications for selectivity
- PMID: 15801829
- PMCID: PMC2896057
- DOI: 10.1021/jm0491703
A comparison of the binding sites of matrix metalloproteinases and tumor necrosis factor-alpha converting enzyme: implications for selectivity
Abstract
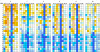
MMPs and TACE (ADAM-17) assume independent, parallel, or opposite pathological roles in cancer, arthritis, and several other diseases. For therapeutic purposes, selective inhibition of individual MMPs and TACE is required in most cases due to distinct roles in diseases and the need to preserve activities in normal states. Toward this goal, we compared force-field interaction energies of five ubiquitous inhibitor atoms with flexible binding sites of 24 known human MMPs and TACE. The results indicate that MMPs 1-3, 10, 11, 13, 16, and 17 have at least one subsite very similar to TACE. S3 subsite is the best target for development of specific TACE inhibitors. Specific binding to TACE compared to most MMPs is promoted by placing a negatively charged ligand part at the bottom of S2 subsite, at the entrance of S1' subsite, or the part of S3' subsite that is close to catalytic zinc. Numerous other clues, consistent with available experimental data, are provided for design of selective inhibitors.
Figures

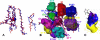


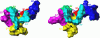
References
-
- Sternlicht MD, Coussens LM, Vu TH, Werb Z. Biology and regulation of the matrix metalloproteinases. In: Clendeninn NJ, Appelt K, editors. Matrix Metalloproteinase Inhibitors in Cancer Therapy. Humana Press; Totowa, NJ: 2000. pp. 1–37.
-
- Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: An overview. Mol. Cell. Biochem. 2003;253:269–285. - PubMed
-
- Stahl M, Taroni C, Schneider G, Hoffmann F. Mapping of protein surface cavities and prediction of enzyme class by a self-organizing neural network. Protein Eng. 2000;13:83–88. - PubMed
-
- Blobel CP. Functional and biochemical characterization of ADAMs and their predicted role in protein ectodomain shedding. Inflamm. Res. 2002;51:83–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

