pRb-Independent growth arrest and transcriptional regulation of E2F target genes
- PMID: 15802019
- PMCID: PMC1501127
- DOI: 10.1593/neo.04394
pRb-Independent growth arrest and transcriptional regulation of E2F target genes
Abstract
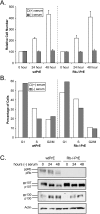
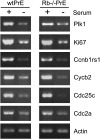
The retinoblastoma tumor suppressor (pRb) has traditionally been studied as a negative regulator of cell cycle progression through its interactions with the E2F family of transcription factors. Utilizing prostate epithelial cell lines established from Rb+/+ and Rb-/- prostate tissues, we previously demonstrated that Rb-/- epithelial cells were not transformed and retained the ability to differentiate in vivo despite the lack of pRb. To further study the effects of pRb loss in an epithelial cell population, we utilized oligonucleotide microarrays to identify any pRb-dependent transcriptional regulation during serum depletion-induced growth arrest. These studies identified 120 unique transcripts regulated by growth arrest in Rb+/+ cells. In these wild-type cells, the majority (80%) of altered transcripts were downregulated, including 40 previously identified E2F target genes. Although the transcriptional repression of E2F target genes is characteristic of pRb pocket protein family activity, further analysis revealed that, compared to Rb+/+ cells, Rb-/- cells exhibited a nearly identical response for all transcripts including those of E2F target genes. These findings demonstrate that pRb is not strictly required for the vast majority of transcriptional alterations associated with growth arrest.
Figures



Similar articles
-
RBP1 induces growth arrest by repression of E2F-dependent transcription.Oncogene. 1999 Mar 25;18(12):2091-100. doi: 10.1038/sj.onc.1202520. Oncogene. 1999. PMID: 10321733
-
Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway.Cancer Res. 2005 May 1;65(9):3624-32. doi: 10.1158/0008-5472.CAN-04-2158. Cancer Res. 2005. PMID: 15867357
-
pRb inactivation in senescent cells leads to an E2F-dependent apoptosis requiring p73.Mol Cancer Res. 2003 Aug;1(10):716-28. Mol Cancer Res. 2003. PMID: 12939397
-
pRB, p107 and the regulation of the E2F transcription factor.J Cell Sci Suppl. 1994;18:81-7. doi: 10.1242/jcs.1994.supplement_18.12. J Cell Sci Suppl. 1994. PMID: 7883798 Review.
-
Activity of the retinoblastoma family proteins, pRB, p107, and p130, during cellular proliferation and differentiation.Crit Rev Biochem Mol Biol. 1996 Jun;31(3):237-71. doi: 10.3109/10409239609106585. Crit Rev Biochem Mol Biol. 1996. PMID: 8817077 Review.
Cited by
-
Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells.Cancer Res. 2007 Jul 1;67(13):6192-203. doi: 10.1158/0008-5472.CAN-06-4424. Cancer Res. 2007. PMID: 17616676 Free PMC article.
-
A review of the past, present, and future directions of neoplasia.Neoplasia. 2005 Dec;7(12):1039-46. doi: 10.1593/neo.05793. Neoplasia. 2005. PMID: 16354585 Free PMC article. Review. No abstract available.
-
Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis.PLoS Biol. 2008 Aug 12;6(8):e194. doi: 10.1371/journal.pbio.0060194. PLoS Biol. 2008. PMID: 18700816 Free PMC article.
-
High levels of global genome methylation in patients with retinoblastoma.Oncol Lett. 2020 Jul;20(1):715-723. doi: 10.3892/ol.2020.11613. Epub 2020 May 13. Oncol Lett. 2020. PMID: 32565997 Free PMC article.
-
Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts.Neoplasia. 2008 Apr;10(4):303-13. doi: 10.1593/neo.07834. Neoplasia. 2008. PMID: 18392140 Free PMC article.
References
-
- Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. - PubMed
-
- Chen PL, Scully P, Shew JY, Wang JY, Lee WH. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. - PubMed
-
- DeCaprio JA, Ludlow JW, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang CM, Livingston DM. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. - PubMed
-
- Akiyama T, Toyoshima K. Marked alteration in phosphorylation of the RB protein during differentiation of human promyelocytic HL60 cells. Oncogene. 1990;5:179–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
