Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites
- PMID: 15802467
- PMCID: PMC1087937
- DOI: 10.1073/pnas.0501646102
Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites
Erratum in
- Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):6238
Abstract
To investigate retroviral integration targeting on a nucleotide scale, we examined the base frequencies directly surrounding cloned in vivo HIV-1, murine leukemia virus, and avian sarcoma/leukosis virus integrations. Base preferences of up to 2-fold the expected frequencies were found for three viruses, representing P values down to <10(-100) and defining what appear to be preferred integration sequences. Offset symmetry reflecting the topology of the integration reaction was found for HIV-1 and avian sarcoma/leukosis virus but not murine leukemia virus, suggesting fundamental differences in the way different retroviral integration complexes interact with host-cell DNA.
Figures
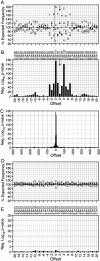
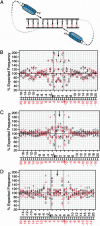
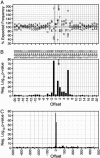
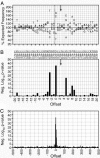
Comment in
-
Symmetrical recognition of cellular DNA target sequences during retroviral integration.Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):5903-4. doi: 10.1073/pnas.0502045102. Epub 2005 Apr 19. Proc Natl Acad Sci U S A. 2005. PMID: 15840713 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

