A high-frequency null mutant of an odorant-binding protein gene, Obp57e, in Drosophila melanogaster
- PMID: 15802511
- PMCID: PMC1450408
- DOI: 10.1534/genetics.104.036483
A high-frequency null mutant of an odorant-binding protein gene, Obp57e, in Drosophila melanogaster
Abstract
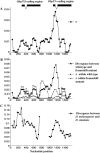
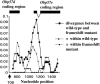
We have found a null mutant of an odorant-binding protein, Obp57e, in Drosophila melanogaster. This frameshift mutation, which is a 10-bp deletion in the coding region, is at a high frequency in the Kyoto population and is also present in Taiwan and Africa. We have sequenced a 1.5-kb region including the tandemly duplicated gene, Obp57d, from 16 inbred lines sampled in Kyoto, Japan. The analyses showed a peak of nucleotide diversity and strong linkage disequilibrium around this mutation. This pattern suggests an elevated mutation rate or an influence of balancing selection in this region. The level of nucleotide divergence between D. melanogaster and D. simulans does not support the former possibility. Thus, this presence/absence polymorphism may be due to balancing selection, which takes advantage of the relatively weak functional constraint in members of a large gene family. In addition, the Obp57d gene region showed an excess of high-frequency-derived mutants that is consistent with a pattern predicted under positive natural selection.
Figures




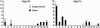
Similar articles
-
Evolution of expression patterns of two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila.Gene. 2010 Nov 1;467(1-2):25-34. doi: 10.1016/j.gene.2010.07.006. Epub 2010 Jul 15. Gene. 2010. PMID: 20637846
-
Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster.Mol Biol Evol. 2009 Jan;26(1):61-70. doi: 10.1093/molbev/msn227. Epub 2008 Oct 14. Mol Biol Evol. 2009. PMID: 18922763
-
Behavioral analyses of mutants for two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila melanogaster.Genes Genet Syst. 2008 Jun;83(3):257-64. doi: 10.1266/ggs.83.257. Genes Genet Syst. 2008. PMID: 18670137
-
[Architecture of the X chromosome, expression of LIM kinase 1, and recombination in the agnostic mutants of Drosophila: a model of human Williams syndrome].Genetika. 2004 Jun;40(6):749-69. Genetika. 2004. PMID: 15341266 Review. Russian.
-
[Allelic exclusion of the odorant receptor genes].Tanpakushitsu Kakusan Koso. 2004 Aug;49(10):1403-12. Tanpakushitsu Kakusan Koso. 2004. PMID: 15346891 Review. Japanese. No abstract available.
Cited by
-
Cis- and Trans-regulatory Effects on Gene Expression in a Natural Population of Drosophila melanogaster.Genetics. 2017 Aug;206(4):2139-2148. doi: 10.1534/genetics.117.201459. Epub 2017 Jun 14. Genetics. 2017. PMID: 28615283 Free PMC article.
-
Cuticular Hydrocarbon Content that Affects Male Mate Preference of Drosophila melanogaster from West Africa.Int J Evol Biol. 2012;2012:278903. doi: 10.1155/2012/278903. Epub 2012 Mar 28. Int J Evol Biol. 2012. PMID: 22536539 Free PMC article.
-
Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia.PLoS Biol. 2007 May;5(5):e118. doi: 10.1371/journal.pbio.0050118. PLoS Biol. 2007. PMID: 17456006 Free PMC article.
-
Rapid evolution of two odorant-binding protein genes, Obp57d and Obp57e, in the Drosophila melanogaster species group.Genetics. 2008 Feb;178(2):1061-72. doi: 10.1534/genetics.107.079046. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245367 Free PMC article.
-
Natural variation in odorant recognition among odorant-binding proteins in Drosophila melanogaster.Genetics. 2010 Mar;184(3):759-67. doi: 10.1534/genetics.109.113340. Epub 2009 Dec 21. Genetics. 2010. PMID: 20026676 Free PMC article.
References
-
- Ayala, F. J., E. S. Balakirev and A. G. Sáez, 2002. Genetic polymorphism at two linked loci, Sod and Est-6, in Drosophila melanogaster. Gene 300: 19–29. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

