Comment on "Oscillations in NF-kappaB signaling control the dynamics of gene expression"
- PMID: 15802586
- PMCID: PMC2821939
- DOI: 10.1126/science.1108198
Comment on "Oscillations in NF-kappaB signaling control the dynamics of gene expression"
Figures
Comment on
-
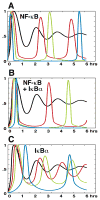
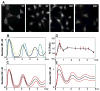
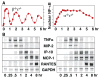
Oscillations in NF-kappaB signaling control the dynamics of gene expression.Science. 2004 Oct 22;306(5696):704-8. doi: 10.1126/science.1099962. Science. 2004. PMID: 15499023
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

