Identification of structural domains involved in astrovirus capsid biology
- PMID: 15802951
- PMCID: PMC1393289
- DOI: 10.1089/vim.2005.18.17
Identification of structural domains involved in astrovirus capsid biology
Abstract
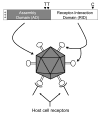
Coat proteins of non-enveloped, icosahedral viruses must perform a variety of functions during their life cycle such as assembly of the coat protein subunits into a closed shell, specific encapsidation of the viral nucleic acid, maturation of the capsid, interaction with host receptors, and disassembly to deliver the genetic information into the newly infected cell. A thorough understanding of the multiple capsid properties at the molecular level is required in order to identify potential targets for antiviral therapy and the prevention of viral disease. The system we have chosen for study is the astrovirus, a family of icosahedral, single-stranded RNA viruses that cause disease in mammals and birds. Very little is known about what regions of the coat protein contribute to the diverse capsid functions. This review will present novel structural predictions for the coat protein sequence of different astrovirus family members. Based on these predictions, we hypothesize that the assembly and RNA packaging functions of the astrovirus coat protein constitutes an individual domain distinct from the determinants required for receptor binding and internalization. Information derived from these structural predictions will serve as an important tool in designing experiments to understand astrovirus biology.
Figures

References
-
- Belloit G, Laveran H, Monroe SS. Capsid protein composition of reference strains and wild isolates of human astroviruses. Virus Res. 1997;49:49–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

