Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism
- PMID: 15803666
- PMCID: PMC2685568
- DOI: 10.1155/2003/162593
Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism
Abstract
Enzymes of the gluconeogenic/glycolytic pathway (the Embden-Meyerhof-Parnas (EMP) pathway), the reductive tricarboxylic acid cycle, the reductive pentose phosphate cycle and the Entner-Doudoroff pathway are widely distributed and are often considered to be central to the origins of metabolism. In particular, several enzymes of the lower portion of the EMP pathway (the so-called trunk pathway), including triosephosphate isomerase (TPI; EC 5.3.1.1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12/13), phosphoglycerate kinase (PGK; EC 2.7.2.3) and enolase (EC 4.2.1.11), are extremely well conserved and universally distributed among the three domains of life. In this paper, the distribution of enzymes of gluconeogenesis/glycolysis in hyperthermophiles--microorganisms that many believe represent the least evolved organisms on the planet--is reviewed. In addition, the phylogenies of the trunk pathway enzymes (TPIs, GAPDHs, PGKs and enolases) are examined. The enzymes catalyzing each of the six-carbon transformations in the upper portion of the EMP pathway, with the possible exception of aldolase, are all derived from multiple gene sequence families. In contrast, single sequence families can account for the archaeal and hyperthermophilic bacterial enzyme activities of the lower portion of the EMP pathway. The universal distribution of the trunk pathway enzymes, in combination with their phylogenies, supports the notion that the EMP pathway evolved in the direction of gluconeogenesis, i.e., from the bottom up.
Figures

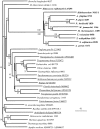
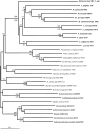

References
-
- Altekar W. Demonstration of some glycolytic activities in Halobacterium halobium and their significance. Indian J. Biochem. Biophys. 1983;20:29–32. - PubMed
-
- Altekar W. A class II fructose-6-phosphate aldolase from a halophilic archaebacterium Haloferax mediterranei . J. Gen. Appl. Microbiol. 1998;44:235–241. - PubMed
-
- Altekar W., Rangaswamy V. Indication of a modified EMP pathway for fructose breakdown in a halophilic archaebacterium. FEMS Microbiol. Lett. 1990;69:139–144.
-
- Altekar W., Rangaswamy V. Degradation of endogenous fructose during catabolism of sucrose and mannitol in halophilic archaebacteria. Arch. Microbiol. 1992;158:356–363.
-
- Altschul S.F., Gish W., Miller W., Myers W., Lipman D.L. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

