Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
- PMID: 15805123
- PMCID: PMC1074309
- DOI: 10.1093/nar/gki337
Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
Abstract
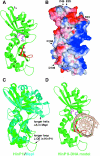
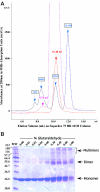
HinP1I, a type II restriction endonuclease, recognizes and cleaves a palindromic tetranucleotide sequence (G/CGC) in double-stranded DNA, producing 2 nt 5' overhanging ends. Here, we report the structure of HinP1I crystallized as one protein monomer in the crystallographic asymmetric unit. HinP1I displays an elongated shape, with a conserved catalytic core domain containing an active-site motif of SDX18QXK and a putative DNA-binding domain. Without significant sequence homology, HinP1I displays striking structural similarity to MspI, an endonuclease that cleaves a similar palindromic DNA sequence (C/CGG) and binds to that sequence crystallographically as a monomer. Almost all the structural elements of MspI can be matched in HinP1I, including both the DNA recognition and catalytic elements. Examining the protein-protein interactions in the crystal lattice, HinP1I could be dimerized through two helices located on the opposite side of the protein to the active site, generating a molecule with two active sites and two DNA-binding surfaces opposite one another on the outer surfaces of the dimer. A possible functional link between this unusual dimerization mode and the tetrameric restriction enzymes is discussed.
Figures





Similar articles
-
Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.Protein Sci. 2005 Oct;14(10):2590-600. doi: 10.1110/ps.051565105. Protein Sci. 2005. PMID: 16195548 Free PMC article.
-
An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site.Structure. 2004 Sep;12(9):1741-7. doi: 10.1016/j.str.2004.07.014. Structure. 2004. PMID: 15341737
-
DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion.Nucleic Acids Res. 2006 Feb 9;34(3):939-48. doi: 10.1093/nar/gkj484. Print 2006. Nucleic Acids Res. 2006. PMID: 16473850 Free PMC article.
-
Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.J Mol Biol. 2008 Dec 12;384(2):489-502. doi: 10.1016/j.jmb.2008.09.033. Epub 2008 Sep 21. J Mol Biol. 2008. PMID: 18835275
-
Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence.EMBO J. 1998 Sep 15;17(18):5466-76. doi: 10.1093/emboj/17.18.5466. EMBO J. 1998. PMID: 9736624 Free PMC article.
Cited by
-
Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.Nucleic Acids Res. 2019 Jan 10;47(1):450-467. doi: 10.1093/nar/gky1059. Nucleic Acids Res. 2019. PMID: 30395313 Free PMC article.
-
The type II restriction endonuclease MvaI has dual specificity.Nucleic Acids Res. 2010 Dec;38(22):8231-8. doi: 10.1093/nar/gkq676. Epub 2010 Aug 6. Nucleic Acids Res. 2010. PMID: 20693529 Free PMC article.
-
Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.Protein Sci. 2005 Oct;14(10):2590-600. doi: 10.1110/ps.051565105. Protein Sci. 2005. PMID: 16195548 Free PMC article.
-
Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI.Nucleic Acids Res. 2007;35(14):4608-18. doi: 10.1093/nar/gkm481. Epub 2007 Jun 22. Nucleic Acids Res. 2007. PMID: 17586812 Free PMC article.
-
BspRI restriction endonuclease: cloning, expression in Escherichia coli and sequential cleavage mechanism.Nucleic Acids Res. 2010 Nov;38(20):7155-66. doi: 10.1093/nar/gkq567. Epub 2010 Jun 29. Nucleic Acids Res. 2010. PMID: 20587501 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

