Cloning and characterization of microRNAs from rice
- PMID: 15805478
- PMCID: PMC1091763
- DOI: 10.1105/tpc.105.031682
Cloning and characterization of microRNAs from rice
Abstract
MicroRNAs (miRNAs) are a growing family of small noncoding RNAs that downregulate gene expression in a sequence-specific manner. The identification of the entire set of miRNAs from a model organism is a critical step toward understanding miRNA-guided gene regulation. Rice (Oryza sativa) and Arabidopsis thaliana, two plant model species with fully sequenced genomes, are representatives of monocotyledonous and dicotyledonous flowering plants, respectively. Thus far, experimental identification of miRNAs in plants has been confined to Arabidopsis. Computational analysis based on conservation with known miRNAs from Arabidopsis has predicted 20 families of miRNAs in rice. To identify miRNAs that are difficult to predict in silico or not conserved in Arabidopsis, we generated three cDNA libraries of small RNAs from rice shoot, root, and inflorescence tissues. We identified 35 miRNAs, of which 14 are new, and these define 13 new families. Thirteen of the new miRNAs are not conserved in Arabidopsis. Four of the new miRNAs are conserved in related monocot species but not in Arabidopsis, which suggests that these may have evolved after the divergence of monocots and dicots. The remaining nine new miRNAs appear to be absent in the known sequences of other plant species. Most of the rice miRNAs are expressed ubiquitously in all tissues examined, whereas a few display tissue-specific expression. We predicted 46 genes as targets of the new rice miRNAs: 16 of these predicted targets encode transcription factors, and other target genes appear to play roles in diverse physiological processes. Four target genes have been experimentally verified by detection of miRNA-mediated mRNA cleavage. Our identification of new miRNAs in rice suggests that these miRNAs may have evolved independently in rice or been lost in other species.
Figures
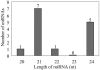






Similar articles
-
Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets.Genome Biol. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. Epub 2004 Aug 31. Genome Biol. 2004. PMID: 15345049 Free PMC article.
-
Conservation and divergence of microRNAs in Populus.BMC Genomics. 2007 Dec 31;8:481. doi: 10.1186/1471-2164-8-481. BMC Genomics. 2007. PMID: 18166134 Free PMC article.
-
Identification of miRNAs and their targets in wheat (Triticum aestivum L.) by EST analysis.Genet Mol Res. 2013 Sep 19;12(3):3793-805. doi: 10.4238/2013.September.19.11. Genet Mol Res. 2013. PMID: 24085441
-
Tomato MicroRNAs and Their Functions.Int J Mol Sci. 2022 Oct 9;23(19):11979. doi: 10.3390/ijms231911979. Int J Mol Sci. 2022. PMID: 36233279 Free PMC article. Review.
-
Conservation and divergence in plant microRNAs.Plant Mol Biol. 2012 Sep;80(1):3-16. doi: 10.1007/s11103-011-9829-2. Epub 2011 Oct 14. Plant Mol Biol. 2012. PMID: 21996939 Review.
Cited by
-
Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells.Nucleic Acids Res. 2012 Nov;40(21):10937-49. doi: 10.1093/nar/gks832. Epub 2012 Sep 10. Nucleic Acids Res. 2012. PMID: 22965126 Free PMC article.
-
Miniature Inverted-repeat Transposable Elements Drive Rapid MicroRNA Diversification in Angiosperms.Mol Biol Evol. 2022 Nov 3;39(11):msac224. doi: 10.1093/molbev/msac224. Mol Biol Evol. 2022. PMID: 36223453 Free PMC article.
-
microRNAs as promising tools for improving stress tolerance in rice.Plant Signal Behav. 2012 Oct 1;7(10):1296-301. doi: 10.4161/psb.21586. Epub 2012 Aug 20. Plant Signal Behav. 2012. PMID: 22902689 Free PMC article. Review.
-
High-Throughput Sequencing Identifies Novel and Conserved Cucumber (Cucumis sativus L.) microRNAs in Response to Cucumber Green Mottle Mosaic Virus Infection.PLoS One. 2015 Jun 15;10(6):e0129002. doi: 10.1371/journal.pone.0129002. eCollection 2015. PLoS One. 2015. PMID: 26076360 Free PMC article.
-
Identification of miRNAs and Their Target Genes Involved in Cucumber Fruit Expansion Using Small RNA and Degradome Sequencing.Biomolecules. 2019 Sep 12;9(9):483. doi: 10.3390/biom9090483. Biomolecules. 2019. PMID: 31547414 Free PMC article.
References
-
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.-H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36, 1282–1290. - PubMed
-
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

