Molecular and functional dissection of the maize B chromosome centromere
- PMID: 15805482
- PMCID: PMC1091764
- DOI: 10.1105/tpc.104.030643
Molecular and functional dissection of the maize B chromosome centromere
Abstract
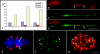
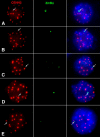
The centromere of the maize (Zea mays) B chromosome contains several megabases of a B-specific repeat (ZmBs), a 156-bp satellite repeat (CentC), and centromere-specific retrotransposons (CRM elements). Here, we demonstrate that only a small fraction of the ZmBs repeats interacts with CENH3, the histone H3 variant specific to centromeres. CentC, which marks the CENH3-associated chromatin in maize A centromeres, is restricted to an approximately 700-kb domain within the larger context of the ZmBs repeats. The breakpoints of five B centromere misdivision derivatives are mapped within this domain. In addition, the fraction of this domain remaining after misdivision correlates well with the quantity of CENH3 on the centromere. Thus, the functional boundaries of the B centromere are mapped to a relatively small CentC- and CRM-rich region that is embedded within multimegabase arrays of the ZmBs repeat. Our results demonstrate that the amount of CENH3 at the B centromere can be varied, but with decreasing amounts, the function of the centromere becomes impaired.
Figures






References
-
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. (2003). Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12, 2711–2721. - PubMed
-
- Carlson, W.R. (1970). Nondisjunction and isochromosome formation in the B chromosome of maize. Chromosoma 30, 356–365.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

