ODORANT1 regulates fragrance biosynthesis in petunia flowers
- PMID: 15805488
- PMCID: PMC1091778
- DOI: 10.1105/tpc.104.028837
ODORANT1 regulates fragrance biosynthesis in petunia flowers
Abstract
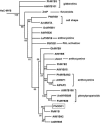
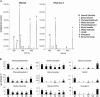
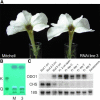
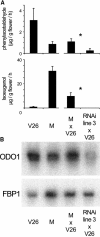
Floral scent is important to plant reproduction because it attracts pollinators to the sexual organs. Therefore, volatile emission is usually tuned to the foraging activity of the pollinators. In Petunia hybrida, volatile benzenoids determine the floral aroma. Although the pathways for benzenoid biosynthesis have been characterized, the enzymes involved are less well understood. How production and emission are regulated is unknown. By targeted transcriptome analyses, we identified ODORANT1 (ODO1), a member of the R2R3-type MYB family, as a candidate for the regulation of volatile benzenoids in Petunia hybrida cv W115 (Mitchell) flowers. These flowers are only fragrant in the evening and at night. Transcript levels of ODO1 increased before the onset of volatile emission and decreased when volatile emission declined. Downregulation of ODO1 in transgenic P. hybrida Mitchell plants strongly reduced volatile benzenoid levels through decreased synthesis of precursors from the shikimate pathway. The transcript levels of several genes in this pathway were reduced by suppression of ODO1 expression. Moreover, ODO1 could activate the promoter of the 5-enol-pyruvylshikimate-3-phosphate synthase gene. Flower pigmentation, which is furnished from the same shikimate precursors, was not influenced because color and scent biosynthesis occur at different developmental stages. Our studies identify ODO1 as a key regulator of floral scent biosynthesis.
Figures

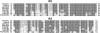

References
-
- Ando, T., Nomura, M., Tsukahara, J., Watanabe, H., Kokubun, H., Tsukamoto, T., Hashimoto, G., Marchesi, E., and Kitching, I.J. (2001). Reproductive isolation in a native population of Petunia sensu Jussieu (Solanaceae). Ann. Bot. (Lond) 88, 403–413.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

