Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana
- PMID: 15805492
- PMCID: PMC1074365
- DOI: 10.1101/gr.3436305
Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana
Abstract
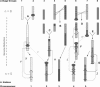
Comparative genome analysis is a powerful tool that can facilitate the reconstruction of the evolutionary history of the genomes of modern-day species. The model plant Arabidopsis thaliana with its n = 5 genome is thought to be derived from an ancestral n = 8 genome. Pairwise comparative genome analyses of A. thaliana with polyploid and diploid Brassicaceae species have suggested that rapid genome evolution, manifested by chromosomal rearrangements and duplications, characterizes the polyploid, but not the diploid, lineages of this family. In this study, we constructed a low-density genetic linkage map of Arabidopsis lyrata ssp. lyrata (A. l. lyrata; n = 8, diploid), the closest known relative of A. thaliana (MRCA approximately 5 Mya), using A. thaliana-specific markers that resolve into the expected eight linkage groups. We then performed comparative Bayesian analyses using raw mapping data from this study and from a Capsella study to infer the number and nature of rearrangements that distinguish the n = 8 genomes of A. l. lyrata and Capsella from the n = 5 genome of A. thaliana. We conclude that there is strong statistical support in favor of the parsimony scenarios of 10 major chromosomal rearrangements separating these n = 8 genomes from A. thaliana. These chromosomal rearrangement events contribute to a rate of chromosomal evolution higher than previously reported in this lineage. We infer that at least seven of these events, common to both sets of data, are responsible for the change in karyotype and underlie genome reduction in A. thaliana.
Figures



References
-
- Acarkan, A., Rossberg, M., Koch, M., and Schmidt, R. 2000. Comparative genome analysis reveals extensive conservation for Arabidopsis thaliana and Capsella rubella. Plant J. 23: 55-62. - PubMed
-
- Al-Shehbaz, I.A. 1973. The biosystematics of the genus Thelypodium (Cruciferae). Contributions to the Gray Herbarium, Harvard University 204: 3-148.
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
