Functional insights from the distribution and role of homopeptide repeat-containing proteins
- PMID: 15805494
- PMCID: PMC1074368
- DOI: 10.1101/gr.3096505
Functional insights from the distribution and role of homopeptide repeat-containing proteins
Abstract
Expansion of "low complex" repeats of amino acids such as glutamine (Poly-Q) is associated with protein misfolding and the development of degenerative diseases such as Huntington's disease. The mechanism by which such regions promote misfolding remains controversial, the function of many repeat-containing proteins (RCPs) remains obscure, and the role (if any) of repeat regions remains to be determined. Here, a Web-accessible database of RCPs is presented. The distribution and evolution of RCPs that contain homopeptide repeats tracts are considered, and the existence of functional patterns investigated. Generally, it is found that while polyamino acid repeats are extremely rare in prokaryotes, several eukaryote putative homologs of prokaryote RCP-involved in important housekeeping processes-retain the repetitive region, suggesting an ancient origin for certain repeats. Within eukarya, the most common uninterrupted amino acid repeats are glutamine, asparagines, and alanine. Interestingly, while poly-Q repeats are found in vertebrates and nonvertebrates, poly-N repeats are only common in more primitive nonvertebrate organisms, such as insects and nematodes. We have assigned function to eukaryote RCPs using Online Mendelian Inheritance in Man (OMIM), the Human Reference Protein Database (HRPD), FlyBase, and Wormpep. Prokaryote RCPs were annotated using BLASTp searches and Gene Ontology. These data reveal that the majority of RCPs are involved in processes that require the assembly of large, multiprotein complexes, such as transcription and signaling.
Figures
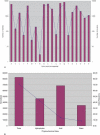
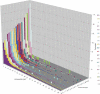
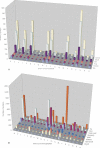

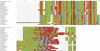


References
-
- Akey, C.W. and Luger, K. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13: 6-14. - PubMed
-
- Alba, M.M., Laskowski, R.A., and Hancock, J.M. 2002. Detecting cryptically simple protein sequences using the SIMPLE algorithm. Bioinformatics 18: 672-678. - PubMed
-
- Barton, G.J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6: 37-40. - PubMed
Web site references
-
- http://repeats.med.monash.edu.au; A database of homopeptide repeats.
-
- http://www.hprd.org/; Human Protein Reference Database.
-
- ftp://ftp.ncbi.nih.gov/blast/db/; NCBI ftp site of available databases.
-
- http://www.ncbi.nlm.nih.gov/omim/; Online Mendelian Inheritance in Man, OMIM. McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), 2000.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
