Diversity of genome structure in Salmonella enterica serovar Typhi populations
- PMID: 15805510
- PMCID: PMC1070368
- DOI: 10.1128/JB.187.8.2638-2650.2005
Diversity of genome structure in Salmonella enterica serovar Typhi populations
Abstract
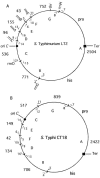
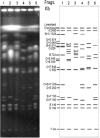
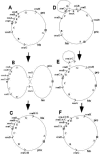
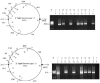
The genomes of most strains of Salmonella and Escherichia coli are highly conserved. In contrast, all 136 wild-type strains of Salmonella enterica serovar Typhi analyzed by partial digestion with I-CeuI (an endonuclease which cuts within the rrn operons) and pulsed-field gel electrophoresis and by PCR have rearrangements due to homologous recombination between the rrn operons leading to inversions and translocations. Recombination between rrn operons in culture is known to be equally frequent in S. enterica serovar Typhi and S. enterica serovar Typhimurium; thus, the recombinants in S. enterica serovar Typhi, but not those in S. enterica serovar Typhimurium, are able to survive in nature. However, even in S. enterica serovar Typhi the need for genome balance and the need for gene dosage impose limits on rearrangements. Of 100 strains of genome types 1 to 6, 72 were only 25.5 kb off genome balance (the relative lengths of the replichores during bidirectional replication from oriC to the termination of replication [Ter]), while 28 strains were less balanced (41 kb off balance), indicating that the survival of the best-balanced strains was greater. In addition, the need for appropriate gene dosage apparently selected against rearrangements which moved genes from their accustomed distance from oriC. Although rearrangements involving the seven rrn operons are very common in S. enterica serovar Typhi, other duplicated regions, such as the 25 IS200 elements, are very rarely involved in rearrangements. Large deletions and insertions in the genome are uncommon, except for deletions of Salmonella pathogenicity island 7 (usually 134 kb) from fragment I-CeuI-G and 40-kb insertions, possibly a prophage, in fragment I-CeuI-E. The phage types were determined, and the origins of the phage types appeared to be independent of the origins of the genome types.
Figures


Similar articles
-
Inversions over the terminus region in Salmonella and Escherichia coli: IS200s as the sites of homologous recombination inverting the chromosome of Salmonella enterica serovar typhi.J Bacteriol. 2002 Nov;184(22):6190-7. doi: 10.1128/JB.184.22.6190-6197.2002. J Bacteriol. 2002. PMID: 12399489 Free PMC article.
-
Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella.FEMS Microbiol Lett. 1998 Jul 15;164(2):275-81. doi: 10.1111/j.1574-6968.1998.tb13098.x. FEMS Microbiol Lett. 1998. PMID: 9682477
-
Rearrangements in the genome of the bacterium Salmonella typhi.Proc Natl Acad Sci U S A. 1995 Feb 14;92(4):1018-22. doi: 10.1073/pnas.92.4.1018. Proc Natl Acad Sci U S A. 1995. PMID: 7862625 Free PMC article.
-
Chromosomal rearrangements in enteric bacteria.Electrophoresis. 1998 Apr;19(4):569-72. doi: 10.1002/elps.1150190417. Electrophoresis. 1998. PMID: 9588803 Review.
-
The genome of Salmonella enterica serovar Typhi.Clin Infect Dis. 2007 Jul 15;45 Suppl 1:S29-33. doi: 10.1086/518143. Clin Infect Dis. 2007. PMID: 17582565 Review.
Cited by
-
Chromosomal rearrangements formed by rrn recombination do not improve replichore balance in host-specific Salmonella enterica serovars.PLoS One. 2010 Oct 19;5(10):e13503. doi: 10.1371/journal.pone.0013503. PLoS One. 2010. PMID: 20976060 Free PMC article.
-
Diversity of Salmonella enterica serovar Typhi strains collected from india using variable number tandem repeat (VNTR)-PCR analysis.Mol Diagn Ther. 2013 Aug;17(4):257-64. doi: 10.1007/s40291-013-0034-7. Mol Diagn Ther. 2013. PMID: 23615945
-
Pathogenicity Factors of Genomic Islands in Intestinal and Extraintestinal Escherichia coli.Front Microbiol. 2020 Sep 25;11:2065. doi: 10.3389/fmicb.2020.02065. eCollection 2020. Front Microbiol. 2020. PMID: 33101219 Free PMC article. Review.
-
Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements.Genome Biol Evol. 2010 Jan 22;2:102-16. doi: 10.1093/gbe/evq003. Genome Biol Evol. 2010. PMID: 20333227 Free PMC article.
-
Insertion Sequence (IS)-Excision Enhancer (IEE)-Mediated IS Excision from the lacZ Gene Restores the Lactose Utilization Defect of Shiga Toxin-Producing Escherichia coli O121:H19 Strains and Is Responsible for Their Delayed Lactose Utilization Phenotype.Appl Environ Microbiol. 2022 Aug 23;88(16):e0076022. doi: 10.1128/aem.00760-22. Epub 2022 Aug 1. Appl Environ Microbiol. 2022. PMID: 35913153 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Anderson, R. P., and J. R. Roth. 1979. Gene duplication in bacteria: alteration of gene dosage by sister chromosome exchanges. Cold Spring Harbor Symp. Quant. Biol. 43:1083-1087. - PubMed
-
- Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

