Protein diversity confers specificity in plasmid segregation
- PMID: 15805511
- PMCID: PMC1070370
- DOI: 10.1128/JB.187.8.2651-2661.2005
Protein diversity confers specificity in plasmid segregation
Abstract
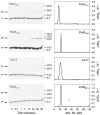
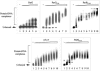
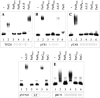
The ParG segregation protein (8.6 kDa) of multidrug resistance plasmid TP228 is a homodimeric DNA-binding factor. The ParG dimer consists of intertwined C-terminal domains that adopt a ribbon-helix-helix architecture and a pair of flexible, unstructured N-terminal tails. A variety of plasmids possess partition loci with similar organizations to that of TP228, but instead of ParG homologs, these plasmids specify a diversity of unrelated, but similarly sized, partition proteins. These include the proteobacterial pTAR, pVT745, and pB171 plasmids. The ParG analogs of these plasmids were characterized in parallel with the ParG homolog encoded by the pseudomonal plasmid pVS1. Like ParG, the four proteins are dimeric. No heterodimerization was detectable in vivo among the proteins nor with the prototypical ParG protein, suggesting that monomer-monomer interactions are specific among the five proteins. Nevertheless, as with ParG, the ParG analogs all possess significant amounts of unordered amino acid residues, potentially highlighting a common structural link among the proteins. Furthermore, the ParG analogs bind specifically to the DNA regions located upstream of their homologous parF-like genes. These nucleoprotein interactions are largely restricted to cognate protein-DNA pairs. The results reveal that the partition complexes of these and related plasmids have recruited disparate DNA-binding factors that provide a layer of specificity to the macromolecular interactions that mediate plasmid segregation.
Figures



Similar articles
-
ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure.Mol Microbiol. 2003 Nov;50(4):1141-53. doi: 10.1046/j.1365-2958.2003.03750.x. Mol Microbiol. 2003. PMID: 14622405
-
Architecture of the ParF*ParG protein complex involved in prokaryotic DNA segregation.Mol Microbiol. 2003 Jul;49(2):487-99. doi: 10.1046/j.1365-2958.2003.03564.x. Mol Microbiol. 2003. PMID: 12828644
-
The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression.J Biol Chem. 2005 Aug 5;280(31):28683-91. doi: 10.1074/jbc.M501173200. Epub 2005 Jun 12. J Biol Chem. 2005. PMID: 15951570
-
Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation.Biochem J. 2008 May 15;412(1):1-18. doi: 10.1042/BJ20080359. Biochem J. 2008. PMID: 18426389 Review.
-
Plasmid and chromosome partitioning: surprises from phylogeny.Mol Microbiol. 2000 Aug;37(3):455-66. doi: 10.1046/j.1365-2958.2000.01975.x. Mol Microbiol. 2000. PMID: 10931339 Review.
Cited by
-
Genome Segregation by the Venus Flytrap Mechanism: Probing the Interaction Between the ParF ATPase and the ParG Centromere Binding Protein.Front Mol Biosci. 2020 Jun 16;7:108. doi: 10.3389/fmolb.2020.00108. eCollection 2020. Front Mol Biosci. 2020. PMID: 32613008 Free PMC article.
-
Centromere binding and evolution of chromosomal partition systems in the Burkholderiales.J Bacteriol. 2012 Jul;194(13):3426-36. doi: 10.1128/JB.00041-12. Epub 2012 Apr 20. J Bacteriol. 2012. PMID: 22522899 Free PMC article.
-
Function, expression, specificity, diversity and incompatibility of actinobacteriophage parABS systems.Mol Microbiol. 2016 Aug;101(4):625-44. doi: 10.1111/mmi.13414. Epub 2016 Jun 10. Mol Microbiol. 2016. PMID: 27146086 Free PMC article.
-
Tubulin homolog TubZ in a phage-encoded partition system.Proc Natl Acad Sci U S A. 2012 May 15;109(20):7711-6. doi: 10.1073/pnas.1121546109. Epub 2012 Apr 26. Proc Natl Acad Sci U S A. 2012. PMID: 22538818 Free PMC article.
-
Catching a Walker in the Act-DNA Partitioning by ParA Family of Proteins.Front Microbiol. 2022 May 26;13:856547. doi: 10.3389/fmicb.2022.856547. eCollection 2022. Front Microbiol. 2022. PMID: 35694299 Free PMC article. Review.
References
-
- Alberts, B. 1998. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92:291-294. - PubMed
-
- Austin, S., and K. Nordstrom. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. - PubMed
-
- Barillà, D., and F. Hayes. 2003. Architecture of the ParF-ParG protein complex involved in procaryotic DNA segregation. Mol. Microbiol. 49:487-499. - PubMed
-
- Benos, P. V., A. S. Lapedes, and G. D. Stormo. 2002. Is there a code for protein-DNA recognition? Probab(ilistical)ly… Bioessays 24:466-475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

