Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins
- PMID: 15809312
- PMCID: PMC2171914
- DOI: 10.1083/jcb.200412059
Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins
Abstract
Viral fusion proteins of classes I and II differ radically in their initial structures but refold toward similar conformations upon activation. Do fusion pathways mediated by alphavirus E1 and influenza virus hemagglutinin (HA) that exemplify classes II and I differ to reflect the difference in their initial conformations, or concur to reflect the similarity in the final conformations? Here, we dissected the pathway of low pH-triggered E1-mediated cell-cell fusion by reducing the numbers of activated E1 proteins and by blocking different fusion stages with specific inhibitors. The discovered progression from transient hemifusion to small, and then expanding, fusion pores upon an increase in the number of activated fusion proteins parallels that established for HA-mediated fusion. We conclude that proteins as different as E1 and HA drive fusion through strikingly similar membrane intermediates, with the most energy-intensive stages following rather than preceding hemifusion. We propose that fusion reactions catalyzed by all proteins of both classes follow a similar pathway.
Figures
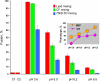
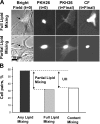


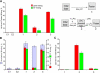
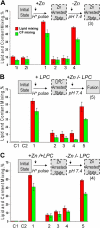

References
-
- Borrego-Diaz, E., M.E. Peeples, R.M. Markosyan, G.B. Melikyan, and F.S. Cohen. 2003. Completion of trimeric hairpin formation of influenza virus hemagglutinin promotes fusion pore opening and enlargement. Virology. 316:234–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

