Transaldolase of Methanocaldococcus jannaschii
- PMID: 15810435
- PMCID: PMC2685571
- DOI: 10.1155/2004/608428
Transaldolase of Methanocaldococcus jannaschii
Abstract
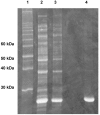
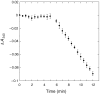
The Methanocaldococcus jannaschii genome contains putative genes for all four nonoxidative pentose phosphate pathway enzymes. Open reading frame (ORF) MJ0960 is a member of the mipB/talC family of 'transaldolase-like' genes, so named because of their similarity to the well-characterized transaldolase B gene family. However, recently, it has been reported that both the mipB and the talC genes from Escherichia coli encode novel enzymes with fructose-6-phosphate aldolase activity, not transaldolase activity (Schürmann and Sprenger 2001). The same study reports that other members of the mipB/talC family appear to encode transaldolases. To confirm the function of MJ0960 and to clarify the presence of a nonoxidative pentose phosphate pathway in M. jannaschii, we have cloned ORF MJ0960 from M. jannaschii genomic DNA and purified the recombinant protein. MJ0960 encodes a transaldolase and displays no fructose-6-phosphate aldolase activity. It etained full activity for 4 h at 80 degrees C, and for 3 weeks at 25 degrees C. Methanocaldococcus jannaschii transaldolase has a maximal velocity (Vmax) of 1.0 +/- 0.2 micromol min(-1) mg(-1) at 25 degrees C, whereas Vmax = 12.0 +/- 0.5 micromol min(-1) mg(-1) at 50 degrees C. Apparent Michaelis constants at 50 degrees C were Km = 0.65 +/- 0.09 mM for fructose-6-phosphate and Km = 27.8 +/- 4.3 microM for erythrose-4-phosphate. When ribose-5-phosphate replaced erythrose-4-phosphate as an aldose acceptor, Vmax decreased twofold, whereas the Km was 150-fold higher. The molecular mass of the active enzyme is 271 +/- 27 kDa as estimated by gel filtration, whereas the predicted monomer size is 23.96 kDa, suggesting that the native form of the protein is probably a decamer. A readily available source of thermophilic pentose phosphate pathway enzymes including transaldolase may have direct application in enzymatic biohydrogen production.
Figures


References
-
- Bruins M.E., Janssen A.E.M., Boom R.M. Thermozymes and their applications: a review of recent literature and patents. Appl. Biochem. Biotechnol. 2001;90:155–186. - PubMed
-
- Brunner N.A., Brinkmann H., Siebers B., Hensel R. NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteustenax . J. Biol. Chem. 1998;273:6149–6156. - PubMed
-
- Choquet C.G., Richards J.C., Patel G.B., Sprott G.D. Ribose biosynthesis in methanogenic bacteria. Arch. Microbiol. 1994;161:481–488.
-
- Cordwell S.J. Microbial genomes and “missing” enzymes: redefining biochemical pathways. Arch. Microbiol. 1999;172:269–279. - PubMed
-
- Haki G.D., Rakshit S.K. Developments in industrially important thermostable enzymes: a review. Bioresour. Technol. 2003;89:17–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

