Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont
- PMID: 15812043
- PMCID: PMC1082560
- DOI: 10.1128/AEM.71.4.2095-2105.2005
Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont
Abstract
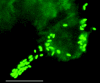
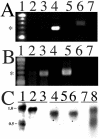
We developed and applied transposon-based transformation vectors for molecular manipulation and analysis of spotted fever group rickettsiae, which are obligate intracellular bacteria that infect ticks and, in some cases, mammals. Using the Epicentre EZ::TN transposon system, we designed transposons for simultaneous expression of a reporter gene and a chloramphenicol acetyltransferase (CAT) resistance marker. Transposomes (transposon-transposase complexes) were electroporated into Rickettsia monacensis, a rickettsial symbiont isolated from the tick Ixodes ricinus. Each transposon contained an expression cassette consisting of the rickettsial ompA promoter and a green fluorescent protein (GFP) reporter gene (GFPuv) or the ompB promoter and a red fluorescent protein reporter gene (DsRed2), followed by the ompA transcription terminator and a second ompA promoter CAT gene cassette. Selection with chloramphenicol gave rise to rickettsial populations with chromosomally integrated single-copy transposons as determined by PCR, Southern blotting, and sequence analysis. Reverse transcription-PCR and Northern blots demonstrated transcription of all three genes. GFPuv transformant rickettsiae exhibited strong fluorescence in individual cells, but DsRed2 transformants did not. Western blots confirmed expression of GFPuv in R. monacensis and in Escherichia coli, but DsRed2 was expressed only in E. coli. The DsRed2 gene, but not the GFPuv gene, contains many GC-rich amino acid codons that are rare in the preferred codon suite of rickettsiae, possibly explaining the failure to express DsRed2 protein in R. monacensis. We demonstrated that our vectors provide a means to study rickettsia-host cell interactions by visualizing GFPuv-fluorescent R. monacensis associated with actin tails in tick host cells.
Figures



Similar articles
-
Rickettsial ompB promoter regulated expression of GFPuv in transformed Rickettsia montanensis.PLoS One. 2010 Jan 29;5(1):e8965. doi: 10.1371/journal.pone.0008965. PLoS One. 2010. PMID: 20126457 Free PMC article.
-
Infection of Ixodes scapularis ticks with Rickettsia monacensis expressing green fluorescent protein: a model system.J Invertebr Pathol. 2007 Mar;94(3):163-74. doi: 10.1016/j.jip.2006.10.003. Epub 2006 Nov 27. J Invertebr Pathol. 2007. PMID: 17125789 Free PMC article.
-
Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis.Appl Environ Microbiol. 2007 Aug;73(15):4984-95. doi: 10.1128/AEM.00988-07. Epub 2007 Jun 15. Appl Environ Microbiol. 2007. PMID: 17575002 Free PMC article.
-
[Emerging rickettsioses].Parassitologia. 2004 Jun;46(1-2):123-6. Parassitologia. 2004. PMID: 15305700 Review. Italian.
-
From genes to proteins: in vitro expression of rickettsial proteins.Ann N Y Acad Sci. 2003 Jun;990:642-52. doi: 10.1111/j.1749-6632.2003.tb07439.x. Ann N Y Acad Sci. 2003. PMID: 12860702 Review.
Cited by
-
Actin-based motility of bacterial pathogens: mechanistic diversity and its impact on virulence.Pathog Dis. 2016 Nov 1;74(8):ftw099. doi: 10.1093/femspd/ftw099. Epub 2016 Sep 20. Pathog Dis. 2016. PMID: 27655913 Free PMC article.
-
Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape.Infect Immun. 2005 Oct;73(10):6668-73. doi: 10.1128/IAI.73.10.6668-6673.2005. Infect Immun. 2005. PMID: 16177343 Free PMC article.
-
Mariner-based transposon mutagenesis of Rickettsia prowazekii.Appl Environ Microbiol. 2007 Oct;73(20):6644-9. doi: 10.1128/AEM.01727-07. Epub 2007 Aug 24. Appl Environ Microbiol. 2007. PMID: 17720821 Free PMC article.
-
Wide dispersal and possible multiple origins of low-copy-number plasmids in rickettsia species associated with blood-feeding arthropods.Appl Environ Microbiol. 2010 Mar;76(6):1718-31. doi: 10.1128/AEM.02988-09. Epub 2010 Jan 22. Appl Environ Microbiol. 2010. PMID: 20097813 Free PMC article.
-
An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum.PLoS Pathog. 2015 Nov 6;11(11):e1005248. doi: 10.1371/journal.ppat.1005248. eCollection 2015. PLoS Pathog. 2015. PMID: 26544981 Free PMC article.
References
-
- Andersson, S. G. E., and P. M. Sharp. 1996. Codon usage and base composition in Rickettsia prowazekii. J. Mol. Evol. 42:525-536. - PubMed
-
- Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. M. Alsmark, R. M. Podowski, A. K. Naslund, A. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. - PubMed
-
- Anonymous. 2001. Living colors DsRed2. Clonetechniques 16:2-3.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

