Impact of human population history on distributions of individual-level genetic distance
- PMID: 15814064
- PMCID: PMC3525116
- DOI: 10.1186/1479-7364-2-1-4
Impact of human population history on distributions of individual-level genetic distance
Abstract
Summaries of human genomic variation shed light on human evolution and provide a framework for biomedical research. Variation is often summarised in terms of one or a few statistics (eg F(ST) and gene diversity). Now that multilocus genotypes for hundreds of autosomal loci are available for thousands of individuals, new approaches are applicable. Recently, trees of individuals and other clustering approaches have demonstrated the power of an individual-focused analysis. We propose analysing the distributions of genetic distances between individuals. Each distribution, or common ancestry profile (CAP), is unique to an individual, and does not require a priori assignment of individuals to populations. Here, we consider a range of models of population history and, using coalescent simulation, reveal the potential insights gained from a set of CAPs. Information lies in the shapes of individual profiles--sometimes captured by variance of individual CAPs--and the variation across profiles. Analysis of short tandem repeat genotype data for over 1,000 individuals from 52 populations is consistent with dramatic differences in population histories across human groups.
Figures
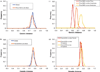

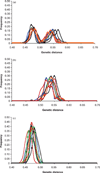


References
-
- Cavalli-Sforza LL, Piazza A, Menozzi P. History and Geography of Human Genes. Princeton University Press, Princeton, NJ; 1994.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

