Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction
- PMID: 15814815
- PMCID: PMC1074725
- DOI: 10.1093/nar/gki348
Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction
Abstract
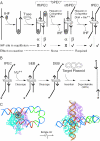
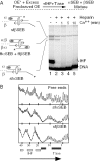
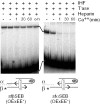
The Tn10 transpososome is a DNA processing machine in which two transposon ends, a transposase dimer and the host protein integration host factor (IHF), are united in an asymmetrical complex. The transitions that occur during one transposition cycle are not limited to chemical cleavage events at the transposon ends, but also involve a reorganization of the protein and DNA components. Here, we demonstrate multiple pathways for Tn10 transposition. We show that one series of events is favored over all others and involves cyclic changes in the affinity of IHF for its binding site. During transpososome assembly, IHF is bound with high affinity. However, the affinity for IHF drops dramatically after cleavage of the first transposon end, leading to IHF ejection and unfolding of the complex. The ejection of IHF promotes cleavage of the second end, which is followed by restoration of the high affinity state which in turn regulates target interactions.
Figures
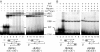


Similar articles
-
The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition.Genes Dev. 2005 Sep 15;19(18):2224-35. doi: 10.1101/gad.1338905. Genes Dev. 2005. PMID: 16166383 Free PMC article.
-
The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms.Nucleic Acids Res. 2003 Oct 15;31(20):5868-76. doi: 10.1093/nar/gkg797. Nucleic Acids Res. 2003. PMID: 14530435 Free PMC article.
-
Transpososome dynamics and regulation in Tn10 transposition.Crit Rev Biochem Mol Biol. 2006 Nov-Dec;41(6):407-24. doi: 10.1080/10409230600987415. Crit Rev Biochem Mol Biol. 2006. PMID: 17092825 Review.
-
DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution.Mol Cell. 2004 Feb 27;13(4):537-47. doi: 10.1016/s1097-2765(04)00052-8. Mol Cell. 2004. PMID: 14992723
-
Tn5 as a model for understanding DNA transposition.Mol Microbiol. 2003 Mar;47(5):1199-206. doi: 10.1046/j.1365-2958.2003.03382.x. Mol Microbiol. 2003. PMID: 12603728 Review.
Cited by
-
Mutation of Tn5 transposase beta-loop residues affects all steps of Tn5 transposition: the role of conformational changes in Tn5 transposition.Biochemistry. 2006 Dec 26;45(51):15552-62. doi: 10.1021/bi061227v. Epub 2006 Dec 5. Biochemistry. 2006. PMID: 17176076 Free PMC article.
-
Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks.Mol Cell Biol. 2009 Nov;29(21):5889-99. doi: 10.1128/MCB.00187-09. Epub 2009 Aug 31. Mol Cell Biol. 2009. PMID: 19720743 Free PMC article.
-
Bacterial stationary-state mutagenesis and Mammalian tumorigenesis as stress-induced cellular adaptations and the role of epigenetics.Curr Genomics. 2006;7(8):481-96. doi: 10.2174/138920206779315764. Curr Genomics. 2006. PMID: 18369407 Free PMC article.
-
A simple topological filter in a eukaryotic transposon as a mechanism to suppress genome instability.Mol Cell Biol. 2011 Jan;31(2):317-27. doi: 10.1128/MCB.01066-10. Epub 2010 Nov 1. Mol Cell Biol. 2011. PMID: 21041479 Free PMC article.
-
The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition.Genes Dev. 2005 Sep 15;19(18):2224-35. doi: 10.1101/gad.1338905. Genes Dev. 2005. PMID: 16166383 Free PMC article.
References
-
- Katinka M.D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. - PubMed
-
- Davis B.M., Waldor M.K. Mobile genetic elements and bacterial pathogenesis. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 1040–1059.
-
- Campbell A. Eubacterial genomes. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington, DC: American Society for Microbiology; 2002. pp. 1024–1039.
-
- Chalmers R., Blot M. Insertion sequences and transposons. In: Charlebois R.L., editor. Organization of the Prokaryotic Genome. Washington, DC: American Society for Microbiology; 1999. pp. 151–169.

