Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis
- PMID: 15814839
- PMCID: PMC1142423
- DOI: 10.1091/mbc.e05-01-0079
Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis
Abstract
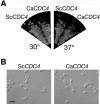
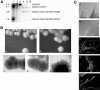
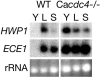
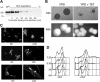
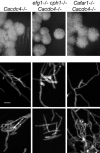
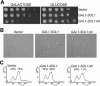
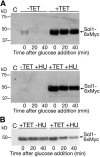
The ability of Candida albicans, a major fungal pathogen, to switch between a yeast form, and a hyphal (mold) form is recognized as being important for the ability of the organism to invade the host and cause disease. We found that a C. albicans mutant deleted for CaCDC4, a homologue of the Saccharomyces cerevisiae F-box protein component of the SCF(CDC4) ubiquitin ligase, is viable and displays constitutive filamentous, mostly hyphal, growth. The phenotype of the Cacdc4-/- mutant suggests that ubiquitin-mediated protein degradation is involved in the regulation of the dimorphic switch of C. albicans and that one or more regulators of the yeast-to-mold switch are among the substrates of SCF(CaCDC4). Epistasis analysis indicates that the Cacdc4-/- phenotype is largely independent of the filamentation-inducing transcription factors Efg1 and Cph1. We identify C. albicans Far1 and Sol1, homologues of the S. cerevisiae SCF(CDC4) substrates Far1 and Sic1, and show that Sol1 is a substrate of C. albicans Cdc4. Neither protein is essential for the hyphal phenotype of the Cacdc4-/- mutant. However, ectopic expression and deletion of SOL1 indicate a role for this gene in C. albicans morphogenesis.
Figures





References
-
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., and Elledge, S. J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

