Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans
- PMID: 15814841
- PMCID: PMC1142435
- DOI: 10.1091/mbc.e05-01-0071
Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans
Abstract
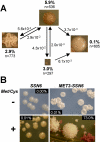
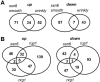
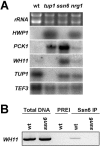
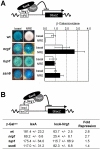
In budding yeast, Tup1 and Ssn6/Cyc8 form a corepressor that regulates a large number of genes. This Tup1-Ssn6 corepressor appears to be conserved from yeast to man. In the pathogenic fungus Candida albicans, Tup1 regulates cellular morphogenesis, phenotypic switching, and metabolism, but the role of Ssn6 remains unclear. We show that there are clear differences in the morphological and invasive phenotypes of C. albicans ssn6 and tup1 mutants. Unlike Tup1, Ssn6 depletion promoted morphological events reminiscent of phenotypic switching rather than filamentous growth. Transcript profiling revealed minimal overlap between the Ssn6 and Tup1 regulons. Hypha-specific genes, which are repressed by Tup1 and Nrg1, were not derepressed in ssn6 cells under the conditions studied. In contrast, the phase specific gene WH11 was derepressed in ssn6 cells, but not in tup1 or nrg1 cells. Hence Ssn6 and Tup1 play distinct roles in C. albicans. Nevertheless, both Ssn6 and Tup1 were required for the Nrg1-mediated repression of an artificial NRE promoter, and lexA-Nrg1 mediated repression in the C. albicans one-hybrid system. These observations are explained in models that are generally consistent with the Tup1-Ssn6 paradigm in budding yeast.
Figures



Similar articles
-
NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans.EMBO J. 2001 Sep 3;20(17):4742-52. doi: 10.1093/emboj/20.17.4742. EMBO J. 2001. PMID: 11532938 Free PMC article.
-
Ssn6, an important factor of morphological conversion and virulence in Candida albicans.Mol Microbiol. 2003 Feb;47(4):1029-43. doi: 10.1046/j.1365-2958.2003.03353.x. Mol Microbiol. 2003. PMID: 12581357
-
Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2.J Mol Biol. 2001 Jun 22;309(5):1007-15. doi: 10.1006/jmbi.2001.4742. J Mol Biol. 2001. PMID: 11399075
-
Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes.Trends Biochem Sci. 2000 Jul;25(7):325-30. doi: 10.1016/s0968-0004(00)01592-9. Trends Biochem Sci. 2000. PMID: 10871883 Review.
-
Candida albicans hyphal initiation and elongation.Trends Microbiol. 2014 Dec;22(12):707-14. doi: 10.1016/j.tim.2014.09.001. Epub 2014 Sep 25. Trends Microbiol. 2014. PMID: 25262420 Free PMC article. Review.
Cited by
-
Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance.PLoS Biol. 2011 Jul;9(7):e1001105. doi: 10.1371/journal.pbio.1001105. Epub 2011 Jul 19. PLoS Biol. 2011. PMID: 21811397 Free PMC article.
-
Ssn6 Defines a New Level of Regulation of White-Opaque Switching in Candida albicans and Is Required For the Stochasticity of the Switch.mBio. 2016 Jan 26;7(1):e01565-15. doi: 10.1128/mBio.01565-15. mBio. 2016. PMID: 26814177 Free PMC article.
-
Morphogenesis in Candida albicans.Annu Rev Microbiol. 2007;61:529-53. doi: 10.1146/annurev.micro.61.080706.093341. Annu Rev Microbiol. 2007. PMID: 17506678 Free PMC article. Review.
-
Regulatory network modelling of iron acquisition by a fungal pathogen in contact with epithelial cells.BMC Syst Biol. 2010 Nov 4;4:148. doi: 10.1186/1752-0509-4-148. BMC Syst Biol. 2010. PMID: 21050438 Free PMC article.
-
Regulation of phenotypic transitions in the fungal pathogen Candida albicans.Virulence. 2012 May 1;3(3):251-61. doi: 10.4161/viru.20010. Epub 2012 May 1. Virulence. 2012. PMID: 22546903 Free PMC article. Review.
References
-
- Braun, B. R., and Johnson, A. D. (1997). Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277, 105-109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

