Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells
- PMID: 15814843
- PMCID: PMC1142430
- DOI: 10.1091/mbc.e05-01-0041
Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells
Abstract
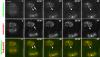
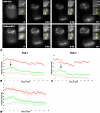
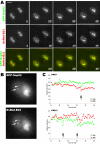
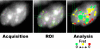
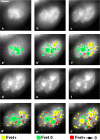
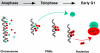
Reorganization of the nuclear machinery after mitosis is a fundamental but poorly understood process. Here, we investigate the recruitment of the nucleolar processing proteins in the nucleolus of living cells at the time of nucleus formation. We question the role of the prenucleolar bodies (PNBs), during migration of the processing proteins from the chromosome periphery to sites of rDNA transcription. Surprisingly, early and late processing proteins pass through the same PNBs as demonstrated by rapid two-color four-dimensional imaging and quantification, whereas a different order of processing protein recruitment into nucleoli is supported by differential sorting. Protein interactions along the recruitment pathway were investigated using a promising time-lapse analysis of fluorescence resonance energy transfer. For the first time, it was possible to detect in living cells the interactions between proteins of the same rRNA processing machinery in nucleoli. Interestingly interactions between such proteins also occur in PNBs but not at the chromosome periphery. The dynamics of these interactions suggests that PNBs are preassembly platforms for rRNA processing complexes.
Figures
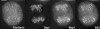
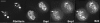

References
-
- Azum-Gélade, M.-C., Noaillac-Depeyre, J., Caizergues-Ferrer, M., and Gas, N. (1994). Cell cycle redistribution of U3 snRNA and fibrillarin. Presence in the cytoplasmic nucleolus remnant and in the prenucleolar bodies at telophase. J. Cell Sci. 107, 463–475. - PubMed
-
- Bastiaens, P. I., and Squire, A. (1999). Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9, 48–52. - PubMed
-
- Bubulya, P. A., Prasanth, K. V., Deerinck, T. J., Gerlich, D., Beaudouin, J., Ellisman, M. H., Ellenberg, J., and Spector, D. L. (2004). Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 167, 51–63. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

