Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis
- PMID: 15814965
- PMCID: PMC1081317
- DOI: 10.1128/JCM.43.4.1552-1563.2005
Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis
Erratum in
- J Clin Microbiol. 2006 Feb;44(2):679
Abstract
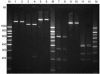
We investigated the genetic relationships of 54 Escherichia coli O103 strains from humans, animals, and meat by molecular typing of housekeeping and virulence genes and by pulsed-field gel electrophoresis (PFGE). Multilocus sequence typing (MLST) of seven housekeeping genes revealed seven profiles, I through VII. MLST profiles I plus III cover 45 Shiga toxin-producing E. coli (STEC) O103:H2 strains from Australia, Canada, France, Germany, and Northern Ireland that are characterized by the intimin (eae) epsilon gene and carry enterohemorrhagic E. coli (EHEC) virulence plasmids. MLST profile II groups five human and animal enteropathogenic E. coli (EPEC) O103:H2 strains that were positive for intimin (eae) beta. Although strains belonging to MLST groups II and I plus III are closely related to each other (92.6% identity), major differences were found in the housekeeping icdA gene and in the virulence-associated genes eae and escD. E. coli O103 strains with MLST patterns IV to VII are genetically distant from MLST I, II, and III strains, as are the non-O103 E. coli strains EDL933 (O157), MG1655 (K-12), and CFT073 (O6). Comparison of MLST results with those of PFGE and virulence typing demonstrated that E. coli O103 STEC and EPEC have recently acquired different virulence genes and DNA rearrangements, causing alterations in their PFGE patterns. PFGE typing was very useful for identification of genetically closely related subgroups among MLST I strains, such as Stx2-producing STEC O103 strains from patients with hemolytic uremic syndrome. Analysis of virulence genes contributed to grouping of E. coli O103 strains into EPEC and STEC. Novel virulence markers, such as efa (EHEC factor for adherence), paa (porcine adherence factor), and cif (cell cycle-inhibiting factor), were found widely associated with E. coli O103 EPEC and STEC strains.
Figures




References
-
- Achtman, M. 2001. A phylogenetic perspective on molecular epidemiology, p. 485-509. In M. Sussmann (ed.), Molecular medical microbiology. Academic Press, San Diego, Calif.
-
- An, H., J. M. Fairbrother, C. Desautels, and J. Harel. 1999. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473:179-184. - PubMed
-
- Badea, L., S. Doughty, L. Nicholls, J. Sloan, R. M. Robins-Browne, and E. L. Hartland. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205-215. - PubMed
-
- Bandelt, H. J., and A. W. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. - PubMed
-
- Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

