Efficient discrimination within a Corynebacterium diphtheriae epidemic clonal group by a novel macroarray-based method
- PMID: 15814981
- PMCID: PMC1081353
- DOI: 10.1128/JCM.43.4.1662-1668.2005
Efficient discrimination within a Corynebacterium diphtheriae epidemic clonal group by a novel macroarray-based method
Erratum in
- J Clin Microbiol. 2005 Jul;43(7):3590
Abstract
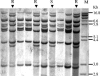
A large diphtheria epidemic in the 1990s in Russia and neighboring countries was caused by a clonal group of closely related Corynebacterium diphtheriae strains (ribotypes Sankt-Peterburg and Rossija). In the recently published complete genome sequence of C. diphtheriae strain NCTC13129, representative of the epidemic clone (A. M. Cerdeno-Tarraga et al., Nucleic Acids Res. 31:6516-6523, 2003), we identified in silico two direct repeat (DR) loci 39 kb downstream and 180 kb upstream of the oriC region, consisting of minisatellite (27- to 36-bp) alternating DRs and variable spacers. We designated these loci DRA and DRB, respectively. A reverse-hybridization macroarray-based method has been developed to study polymorphism (the presence or absence of 21 different spacers) in the larger DRB locus. We name it spoligotyping (spacer oligonucleotide typing), analogously to a similar method of Mycobacterium tuberculosis genotyping. The method was evaluated with 154 clinical strains of the C. diphtheriae epidemic clone from the St. Petersburg area in Russia from 1997 to 2002. By comparison with the international ribotype database (Institut Pasteur, Paris, France), these strains were previously identified as belonging to ribotypes Sankt-Peterburg (n = 79) and Rossija (n = 75). The 154 strains were subdivided into 34 spoligotypes: 14 unique strains and 20 types shared by 2 to 46 strains; the Hunter Gaston discriminatory index (HGDI) was 0.85. DRB locus-based spoligotyping allows fast and efficient discrimination within the C. diphtheriae epidemic clonal group and is applicable to both epidemiological investigations and phylogenetic reconstruction. The results are easy to interpret and can be presented and stored in a user-friendly digital database (Excel file), allowing rapid type determination of new strains.
Figures

Similar articles
-
Corynebacterium diphtheriae spoligotyping based on combined use of two CRISPR loci.Biotechnol J. 2007 Jul;2(7):901-6. doi: 10.1002/biot.200700035. Biotechnol J. 2007. PMID: 17431853
-
Molecular epidemiology of C. diphtheriae strains during different phases of the diphtheria epidemic in Belarus.BMC Infect Dis. 2006 Aug 15;6:129. doi: 10.1186/1471-2334-6-129. BMC Infect Dis. 2006. PMID: 16911772 Free PMC article.
-
Novel macroarray-based method of Corynebacterium diphtheriae genotyping: evaluation in a field study in Belarus.Eur J Clin Microbiol Infect Dis. 2009 Jun;28(6):701-3. doi: 10.1007/s10096-008-0674-4. Epub 2008 Dec 17. Eur J Clin Microbiol Infect Dis. 2009. PMID: 19089478
-
Molecular epidemiology of diphtheria.J Infect Dis. 2000 Feb;181 Suppl 1:S168-77. doi: 10.1086/315556. J Infect Dis. 2000. PMID: 10657209 Review.
-
Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives.Infect Genet Evol. 2009 Jan;9(1):1-15. doi: 10.1016/j.meegid.2008.09.011. Epub 2008 Oct 19. Infect Genet Evol. 2009. PMID: 19007916 Review.
Cited by
-
Whole Genome Sequencing for Surveillance of Diphtheria in Low Incidence Settings.Front Public Health. 2019 Aug 21;7:235. doi: 10.3389/fpubh.2019.00235. eCollection 2019. Front Public Health. 2019. PMID: 31497588 Free PMC article. Review.
-
CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W52-7. doi: 10.1093/nar/gkm360. Epub 2007 May 30. Nucleic Acids Res. 2007. PMID: 17537822 Free PMC article.
-
Genomic analysis of endemic clones of toxigenic and non-toxigenic Corynebacterium diphtheriae in Belarus during and after the major epidemic in 1990s.BMC Genomics. 2017 Nov 13;18(1):873. doi: 10.1186/s12864-017-4276-3. BMC Genomics. 2017. PMID: 29132312 Free PMC article.
-
Evaluation of CRISPR Diversity in the Human Skin Microbiome for Personal Identification.mSystems. 2021 Feb 2;6(1):e01255-20. doi: 10.1128/mSystems.01255-20. mSystems. 2021. PMID: 33531409 Free PMC article.
-
The Role of Gene Editing in Neurodegenerative Diseases.Cell Transplant. 2018 Mar;27(3):364-378. doi: 10.1177/0963689717753378. Epub 2018 May 16. Cell Transplant. 2018. PMID: 29766738 Free PMC article. Review.
References
-
- Canchaya, C., G. Fournous, and H. Brüssow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. - PubMed
-
- Cerdeño-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. G. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dows, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrel, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. - PMC - PubMed
-
- Damian, M., F. Grimont, O. Narvskaya, M. Straut, M. Surdeanu, R. Cojocaru, I. Mokrousov, A. Diaconescu, C. Andronescu, A. Melnic, L. Mutoi, and P. A. D. Grimont. 2002. Study of Corynebacterium diphtheriae strains isolated in Romania, northwestern Russia and the Republic of Moldova. Res. Microbiol. 153:99-106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

