Systematic deletion analysis of fission yeast protein kinases
- PMID: 15821139
- PMCID: PMC1087820
- DOI: 10.1128/EC.4.4.799-813.2005
Systematic deletion analysis of fission yeast protein kinases
Abstract
Eukaryotic protein kinases are key molecules mediating signal transduction that play a pivotal role in the regulation of various biological processes, including cell cycle progression, cellular morphogenesis, development, and cellular response to environmental changes. A total of 106 eukaryotic protein kinase catalytic-domain-containing proteins have been found in the entire fission yeast genome, 44% (or 64%) of which possess orthologues (or nearest homologues) in humans, based on sequence similarity within catalytic domains. Systematic deletion analysis of all putative protein kinase-encoding genes have revealed that 17 out of 106 were essential for viability, including three previously uncharacterized putative protein kinases. Although the remaining 89 protein kinase mutants were able to form colonies under optimal growth conditions, 46% of the mutants exhibited hypersensitivity to at least 1 of the 17 different stress factors tested. Phenotypic assessment of these mutants allowed us to arrange kinases into functional groups. Based on the results of this assay, we propose also the existence of four major signaling pathways that are involved in the response to 17 stresses tested. Microarray analysis demonstrated a significant correlation between the expression signature and growth phenotype of kinase mutants tested. Our complete microarray data sets are available at http://giscompute.gis.a-star.edu.sg/~gisljh/kinome.
Figures

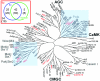





References
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
-
- Beach, D., B. Durkacz, and P. Nurse. 1982. Functionally homologous cell cycle control genes in budding and fission yeast. Nature 300:706-709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

