Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability
- PMID: 15824318
- PMCID: PMC556294
- DOI: 10.1073/pnas.0500617102
Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability
Abstract
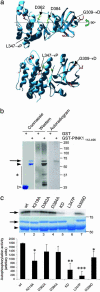
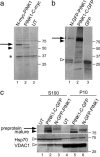
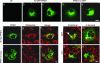
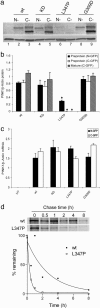
Several mutations in PTEN-induced putative kinase 1 (PINK1) gene have been reported to be associated with recessive parkinsonism. The encoded protein is predicted to be a Ser/Thr protein kinase targeted to mitochondria. In this study, we have investigated the effects of mutations on PINK1 kinase activity in vitro and on expression levels and localization in mammalian cells. We chose to examine two point mutations: G309D, which was originally reported to be stable and properly localized in cells and L347P, which is of interest because it is present at an appreciable carrier frequency in the Philippines. We were able to confirm kinase activity and produce artificial "kinase-dead" mutants that are stable but lack activity. The L347P mutation grossly destabilizes PINK1 and drastically reduces kinase activity, whereas G309D has much more modest effects on these parameters in vitro. This finding is in line with predictions based on homology modeling. We also examined the localization of PINK1 in transfected mammalian cells by using constructs that were tagged with myc or GFP at either end of the protein. These results show that PINK1 is processed at the N terminus in a manner consistent with mitochondrial import, but the mature protein also exists in the cytosol. The physiological relevance of this observation is not yet clear, but it implies that a portion of PINK1 may be exported after processing in the mitochondria.
Figures
References
-
- Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605–608. - PubMed
-
- Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256–259. - PubMed
-
- Hilker, R., Klein, C., Ghaemi, M., Kis, B., Strotmann, T., Ozelius, L. J., Lenz, O., Vieregge, P., Herholz, K., Heiss, W. D. & Pramstaller, P. P. (2001) Ann. Neurol. 49, 367–376. - PubMed
-
- Broussolle, E., Lucking, C. B., Ginovart, N., Pollak, P., Remy, P. & Durr, A. (2000) Neurology 55, 877–879. - PubMed
-
- Scherfler, C., Khan, N. L., Pavese, N., Eunson, L., Graham, E., Lees, A. J., Quinn, N. P., Wood, N. W., Brooks, D. J. & Piccini, P. P. (2004) Brain 127, 1332–1342. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

