Six-helix bundles designed from DNA
- PMID: 15826105
- PMCID: PMC3464188
- DOI: 10.1021/nl050084f
Six-helix bundles designed from DNA
Abstract
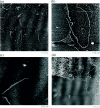
We present a designed cyclic DNA motif that consists of six DNA double helices that are connected to each other at two crossover sites. DNA double helices with 10.5 nucleotide pairs per turn facilitate the programming of DNA double crossover molecules to form hexagonally symmetric arrangements when the crossover points are separated by seven or fourteen nucleotide pairs. We demonstrate by atomic force microscopy well-formed arrays of hexagonal six-helix bundle motifs both in 1D and in 2D.
Figures



Similar articles
-
Three-helix bundle DNA tiles self-assemble into 2D lattice or 1D templates for silver nanowires.Nano Lett. 2005 Apr;5(4):693-6. doi: 10.1021/nl050108i. Nano Lett. 2005. PMID: 15826110
-
Prototyping nanorod control: A DNA double helix sheathed within a DNA six-helix bundle.Chem Biol. 2009 Aug 28;16(8):862-7. doi: 10.1016/j.chembiol.2009.07.008. Chem Biol. 2009. PMID: 19716476
-
Atomic force microscopic measurement of the interdomain angle in symmetric Holliday junctions.Biochemistry. 2002 May 14;41(19):5950-5. doi: 10.1021/bi020001z. Biochemistry. 2002. PMID: 11993988
-
Watson-Crick versus Hoogsteen Base Pairs: Chemical Strategy to Encode and Express Genetic Information in Life.Acc Chem Res. 2021 May 4;54(9):2110-2120. doi: 10.1021/acs.accounts.0c00734. Epub 2021 Feb 16. Acc Chem Res. 2021. PMID: 33591181 Review.
-
An overview of structural DNA nanotechnology.Mol Biotechnol. 2007 Nov;37(3):246-57. doi: 10.1007/s12033-007-0059-4. Epub 2007 Jul 12. Mol Biotechnol. 2007. PMID: 17952671 Free PMC article. Review.
Cited by
-
Multi-micron crisscross structures grown from DNA-origami slats.Nat Nanotechnol. 2023 Mar;18(3):281-289. doi: 10.1038/s41565-022-01283-1. Epub 2022 Dec 21. Nat Nanotechnol. 2023. PMID: 36543881 Free PMC article.
-
Metallic nanoparticles used to estimate the structural integrity of DNA motifs.Biophys J. 2008 Oct;95(7):3340-8. doi: 10.1529/biophysj.108.138479. Epub 2008 Jul 11. Biophys J. 2008. PMID: 18621817 Free PMC article.
-
The biological applications of DNA nanomaterials: current challenges and future directions.Signal Transduct Target Ther. 2021 Oct 8;6(1):351. doi: 10.1038/s41392-021-00727-9. Signal Transduct Target Ther. 2021. PMID: 34620843 Free PMC article. Review.
-
Spatially-interactive biomolecular networks organized by nucleic acid nanostructures.Acc Chem Res. 2012 Aug 21;45(8):1215-26. doi: 10.1021/ar200295q. Epub 2012 May 29. Acc Chem Res. 2012. PMID: 22642503 Free PMC article. Review.
-
Synthesis of DNA Origami Scaffolds: Current and Emerging Strategies.Molecules. 2020 Jul 26;25(15):3386. doi: 10.3390/molecules25153386. Molecules. 2020. PMID: 32722650 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

