Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor
- PMID: 15826239
- PMCID: PMC1180731
- DOI: 10.1042/BJ20050063
Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor
Abstract
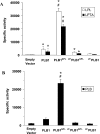
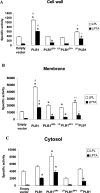
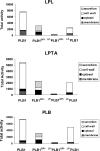
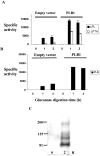
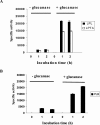
The secreted, multifunctional enzyme PLB1 (phospholipase B1 protein encoded by the PLB1 gene) is a virulence determinant of the pathogenic fungus Cryptococcus neoformans, but the mechanism of its secretion is unknown. The cryptococcal PLB1 gene encodes putative, N-terminal LP (leader peptide) and C-terminal GPI (glycosylphosphatidylinositol) anchor attachment motifs, suggesting that PLB1 is GPI-anchored before secretion. To investigate the role of these motifs in PLB1 secretion, four cDNA constructs were created encoding the full-length construct (PLB1) and three truncated versions without the LP and/or the GPI anchor attachment motifs [(LP-)PLB1 (PLB1 expressed without the LP consensus motif), (LP-)PLB1(GPI-) (PLB1 expressed without the LP and GPI consensus motifs) and PLB1(GPI-) (PLB1 expressed without the GPI anchor attachment motif) respectively]. The constructs were ligated into pYES2, and galactose-induced expression was achieved in Saccharomyces cerevisiae. The LP was essential for secretion of the PLB1 protein and its three activities (PLB, lysophospholipase and lysophospholipase transacylase). Deletion of the GPI motif to create PLB1(GPI-) resulted in a redistribution of activity from the cell wall and membranes to the secreted and cytosolic fractions, with 36-54% of the total activity being secreted as compared with <5% for PLB1. PLB1 produced the maximum cell-associated activity (>2-fold more than that for PLB1(GPI-)), with 75-86% of this in the cell-wall fraction, 6-19% in the membrane fraction and 3-7% in the cytosolic fraction. Cell-wall localization was confirmed by release of activity with beta-glucanase in both S. cerevisiae recombinants and wild-type C. neoformans. The dominant location of PLB1 in the cell wall via GPI anchoring may permit immediate release of the enzyme in response to changing environmental conditions and may represent part of a novel mechanism for regulating the secretion of a fungal virulence determinant.
Figures


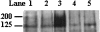
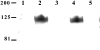
References
-
- Cox G. M., McDade H. C., Chen S. C. A., Tucker S. C., Gottfredsson M., Wright L. C., Sorrell T. C., Leidich S. D., Casadevall A., Ghannoum M. A., et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 2001;39:166–175. - PubMed
-
- Leidich S. D., Ibrahim A. S., Fu Y., Koul A., Jessup C., Vitullo J., Fonzi W., Mirbod F., Nakashima S., Nozawa Y., et al. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 1998;273:26078–26086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

