'PACLIMS': a component LIM system for high-throughput functional genomic analysis
- PMID: 15826298
- PMCID: PMC1090558
- DOI: 10.1186/1471-2105-6-94
'PACLIMS': a component LIM system for high-throughput functional genomic analysis
Abstract
Background: Recent advances in sequencing techniques leading to cost reduction have resulted in the generation of a growing number of sequenced eukaryotic genomes. Computational tools greatly assist in defining open reading frames and assigning tentative annotations. However, gene functions cannot be asserted without biological support through, among other things, mutational analysis. In taking a genome-wide approach to functionally annotate an entire organism, in this application the approximately 11,000 predicted genes in the rice blast fungus (Magnaporthe grisea), an effective platform for tracking and storing both the biological materials created and the data produced across several participating institutions was required.
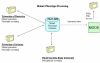
Results: The platform designed, named PACLIMS, was built to support our high throughput pipeline for generating 50,000 random insertion mutants of Magnaporthe grisea. To be a useful tool for materials and data tracking and storage, PACLIMS was designed to be simple to use, modifiable to accommodate refinement of research protocols, and cost-efficient. Data entry into PACLIMS was simplified through the use of barcodes and scanners, thus reducing the potential human error, time constraints, and labor. This platform was designed in concert with our experimental protocol so that it leads the researchers through each step of the process from mutant generation through phenotypic assays, thus ensuring that every mutant produced is handled in an identical manner and all necessary data is captured.
Conclusion: Many sequenced eukaryotes have reached the point where computational analyses are no longer sufficient and require biological support for their predicted genes. Consequently, there is an increasing need for platforms that support high throughput genome-wide mutational analyses. While PACLIMS was designed specifically for this project, the source and ideas present in its implementation can be used as a model for other high throughput mutational endeavors.
Figures



References
-
- Dufresne M, Bailey JA, Dron M, Langin T. clk1, a serine/threonine protein kinase-encoding gene, is involved in pathogenicity of Colletotrichum lindemuthianum on common bean. MPMI. 1998;11:99–108. - PubMed
-
- Sweigard JA, Carroll AM, Farrall L, Chumley FG, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. MPMI. 1998;11:404–412. - PubMed
-
- Balhadère PV, Foster AJ, Talbot NJ. Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea by insertional mutagenesis. Mol Plant Microbe Interact. 1999;12:129–142.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

