Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme
- PMID: 15827123
- PMCID: PMC556127
- DOI: 10.1073/pnas.0402870102
Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme
Abstract
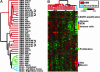
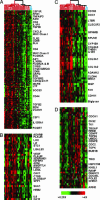
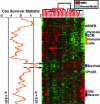
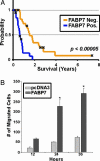
Glioblastoma multiforme (GBM) is the most common form of malignant glioma, characterized by genetic instability, intratumoral histopathological variability, and unpredictable clinical behavior. We investigated global gene expression in surgical samples of brain tumors. Gene expression profiling revealed large differences between normal brain samples and tumor tissues and between GBMs and lower-grade oligodendroglial tumors. Extensive differences in gene expression were found among GBMs, particularly in genes involved in angiogenesis, immune cell infiltration, and extracellular matrix remodeling. We found that the gene expression patterns in paired specimens from the same GBM invariably were more closely related to each other than to any other tumor, even when the paired specimens had strikingly divergent histologies. Survival analyses revealed a set of approximately 70 genes more highly expressed in rapidly progressing tumors that stratified GBMs into two groups that differed by >4-fold in median duration of survival. We further investigated one gene from the group, FABP7, and confirmed its association with survival in two unrelated cohorts totaling 105 patients. Expression of FABP7 enhanced the motility of glioma-derived cells in vitro. Our analyses thus identify and validate a prognostic marker of both biologic and clinical significance and provide a series of putative markers for additional evaluation.
Figures

References
-
- Lacroix, M., Abi-Said, D., Fourney, D. R., Gokaslan, Z. L., Shi, W., DeMonte, F., Lang, F. F., McCutcheon, I. E., Hassenbusch, S. J., Holland, E., et al. (2001) J. Neurosurg. 95, 190–198. - PubMed
-
- Kleihues, P., Louis, D. N., Scheithauer, B. W., Rorke, L. B., Reifenberger, G., Burger, P. C. & Cavenee, W. K. (2002) J. Neuropathol. Exp. Neurol. 61, 215–225; discussion 226–229. - PubMed
-
- Nagane, M., Huang, H. J. & Cavenee, W. K. (1997) Curr. Opin. Oncol. 9, 215–222. - PubMed
-
- Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. - PubMed
-
- Perou, C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

