Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus
- PMID: 15827143
- PMCID: PMC1082772
- DOI: 10.1128/JVI.79.9.5288-5295.2005
Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus
Abstract
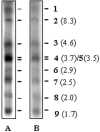
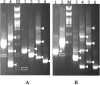
The expression of the genomic information of severe acute respiratory syndrome coronavirus (SARS CoV) involves synthesis of a nested set of subgenomic RNAs (sgRNAs) by discontinuous transcription. In SARS CoV-infected cells, 10 sgRNAs, including 2 novel ones, were identified, which were predicted to be functional in the expression of 12 open reading frames located in the 3' one-third of the genome. Surprisingly, one new sgRNA could lead to production of a truncated spike protein. Sequence analysis of the leader-body fusion sites of each sgRNA showed that the junction sequences and the corresponding transcription-regulatory sequence (TRS) are unique for each species of sgRNA and are consistent after virus passages. For the two novel sgRNAs, each used a variant of the TRS that has one nucleotide mismatch in the conserved hexanucleotide core (ACGAAC) in the TRS. Coexistence of both plus and minus strands of SARS CoV sgRNAs and evidence for derivation of the sgRNA core sequence from the body core sequence favor the model of discontinuous transcription during minus-strand synthesis. Moreover, one rare species of sgRNA has the junction sequence AAA, indicating that its transcription could result from a noncanonical transcription signal. Taken together, these results provide more insight into the molecular mechanisms of genome expression and subgenomic transcription of SARS CoV.
Figures



References
-
- Callebaut, P., I. Correa, M. Pensaert, G. Jimenez, and L. Enjuanes. 1988. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 69:1725-1730. - PubMed
-
- Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

