Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling
- PMID: 15829602
- PMCID: PMC1091762
- DOI: 10.1105/tpc.105.030981
Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling
Abstract
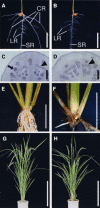
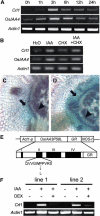
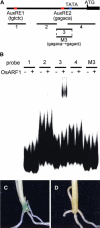
Although the importance of auxin in root development is well known, the molecular mechanisms involved are still unknown. We characterized a rice (Oryza sativa) mutant defective in crown root formation, crown rootless1 (crl1). The crl1 mutant showed additional auxin-related abnormal phenotypic traits in the roots, such as decreased lateral root number, auxin insensitivity in lateral root formation, and impaired root gravitropism, whereas no abnormal phenotypic traits were observed in aboveground organs. Expression of Crl1, which encodes a member of the plant-specific ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES protein family, was localized in tissues where crown and lateral roots are initiated and overlapped with beta-glucuronidase staining controlled by the DR5 promoter. Exogenous auxin treatment induced Crl1 expression without de novo protein biosynthesis, and this induction required the degradation of AUXIN/INDOLE-3-ACETIC ACID proteins. Crl1 contains two putative auxin response elements (AuxREs) in its promoter region. The proximal AuxRE specifically interacted with a rice AUXIN RESPONSE FACTOR (ARF) and acted as a cis-motif for Crl1 expression. We conclude that Crl1 encodes a positive regulator for crown and lateral root formation and that its expression is directly regulated by an ARF in the auxin signaling pathway.
Figures





References
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. - PubMed
-
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8, 165–171. - PubMed
-
- Casson, S.A., and Lindsey, K. (2003). Genes and signaling in root development. New Phytol. 158, 11–38.
-
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. - PubMed
-
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414, 271–276. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

