Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis
- PMID: 15829606
- PMCID: PMC1091768
- DOI: 10.1105/tpc.104.030155
Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis
Abstract
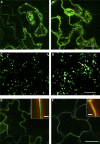
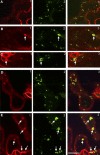
In the absence of cell migration, plant architecture is largely determined by the direction and extent of cell expansion during development. In this report, we show that very-long-chain fatty acid (VLCFA) synthesis plays an essential role in cell expansion. The Arabidopsis thaliana eceriferum10 (cer10) mutants exhibit severe morphological abnormalities and reduced size of aerial organs. These mutants are disrupted in the At3g55360 gene, previously identified as a gene coding for enoyl-CoA reductase (ECR), an enzyme required for VLCFA synthesis. The absence of ECR activity results in a reduction of cuticular wax load and affects VLCFA composition of seed triacylglycerols and sphingolipids, demonstrating in planta that ECR is involved in all VLCFA elongation reactions in Arabidopsis. Epidermal and seed-specific silencing of ECR activity resulted in a reduction of cuticular wax load and the VLCFA content of seed triacylglycerols, respectively, with no effects on plant morphogenesis, suggesting that the developmental phenotypes arise from abnormal sphingolipid composition. Cellular analysis revealed aberrant endocytic membrane traffic and defective cell expansion underlying the morphological defects of cer10 mutants.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

