Characterization of the protease domain of Rice tungro bacilliform virus responsible for the processing of the capsid protein from the polyprotein
- PMID: 15831103
- PMCID: PMC1087892
- DOI: 10.1186/1743-422X-2-33
Characterization of the protease domain of Rice tungro bacilliform virus responsible for the processing of the capsid protein from the polyprotein
Abstract
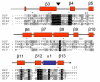
Background: Rice tungro bacilliform virus (RTBV) is a pararetrovirus, and a member of the family Caulimoviridae in the genus Badnavirus. RTBV has a long open reading frame that encodes a large polyprotein (P3). Pararetroviruses show similarities with retroviruses in molecular organization and replication. P3 contains a putative movement protein (MP), the capsid protein (CP), the aspartate protease (PR) and the reverse transcriptase (RT) with a ribonuclease H activity. PR is a member of the cluster of retroviral proteases and serves to proteolytically process P3. Previous work established the N- and C-terminal amino acid sequences of CP and RT, processing of RT by PR, and estimated the molecular mass of PR by western blot assays.
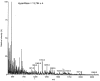
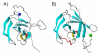
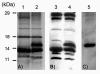
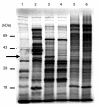
Results: A molecular mass of a protein that was associated with virions was determined by in-line HPLC electrospray ionization mass spectral analysis. Comparison with retroviral proteases amino acid sequences allowed the characterization of a putative protease domain in this protein. Structural modelling revealed strong resemblance with retroviral proteases, with overall folds surrounding the active site being well conserved. Expression in E. coli of putative domain was affected by the presence or absence of the active site in the construct. Analysis of processing of CP by PR, using pulse chase labelling experiments, demonstrated that the 37 kDa capsid protein was dependent on the presence of the protease in the constructs.
Conclusion: The findings suggest the characterization of the RTBV protease domain. Sequence analysis, structural modelling, in vitro expression studies are evidence to consider the putative domain as being the protease domain. Analysis of expression of different peptides corresponding to various domains of P3 suggests a processing of CP by PR. This work clarifies the organization of the RTBV polyprotein, and its processing by the RTBV protease.
Figures


Similar articles
-
Rice tungro bacilliform virus encodes reverse transcriptase, DNA polymerase, and ribonuclease H activities.Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2654-8. doi: 10.1073/pnas.91.7.2654. Proc Natl Acad Sci U S A. 1994. PMID: 7511816 Free PMC article.
-
Rice tungro bacilliform virus open reading frame 3 encodes a single 37-kDa coat protein.Virology. 1999 Jan 20;253(2):319-26. doi: 10.1006/viro.1998.9519. Virology. 1999. PMID: 9918890
-
Analysis of the proteolytic processing and activation of the rice tungro bacilliform virus reverse transcriptase.Virology. 1995 Apr 1;208(1):207-14. doi: 10.1006/viro.1995.1144. Virology. 1995. PMID: 11831702
-
Structural and Functional Aspects of Foamy Virus Protease-Reverse Transcriptase.Viruses. 2019 Jul 2;11(7):598. doi: 10.3390/v11070598. Viruses. 2019. PMID: 31269675 Free PMC article. Review.
-
C-terminal domains of bacterial proteases: structure, function and the biotechnological applications.J Appl Microbiol. 2017 Jan;122(1):12-22. doi: 10.1111/jam.13317. Epub 2016 Nov 10. J Appl Microbiol. 2017. PMID: 27709728 Review.
Cited by
-
Ortervirales: New Virus Order Unifying Five Families of Reverse-Transcribing Viruses.J Virol. 2018 May 29;92(12):e00515-18. doi: 10.1128/JVI.00515-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618642 Free PMC article. No abstract available.
-
Bacilliform DNA-containing plant viruses in the tropics: commonalities within a genetically diverse group.Mol Plant Pathol. 2013 Oct;14(8):759-71. doi: 10.1111/mpp.12046. Epub 2013 Jun 13. Mol Plant Pathol. 2013. PMID: 23763585 Free PMC article. Review.
-
Phylogenetic analysis of Rice tungro bacilliform virus ORFs revealed strong correlation between evolution and geographical distribution.Virus Genes. 2011 Dec;43(3):398-408. doi: 10.1007/s11262-011-0647-z. Epub 2011 Jul 28. Virus Genes. 2011. PMID: 21796436
-
Further support of genetic conservation in Indian isolates of Rice tungro bacilliform virus by sequence analysis of an isolate from North-Western India.Virus Genes. 2013 Apr;46(2):387-91. doi: 10.1007/s11262-012-0857-z. Epub 2012 Nov 30. Virus Genes. 2013. PMID: 23197138
-
Plant Viral Proteases: Beyond the Role of Peptide Cutters.Front Plant Sci. 2018 May 17;9:666. doi: 10.3389/fpls.2018.00666. eCollection 2018. Front Plant Sci. 2018. PMID: 29868107 Free PMC article. Review.
References
-
- Hull R, Geering A, Harper G, Lockhart BE, Schoelz JE. Caulimoviridae. In: Fauquet C.M. MMAMJDUBLA, editor. Virus Taxonomy, VIIIth Report of the ICTV. London , Elsevier/Academic Press; 2005. pp. 385–396.
-
- Takatsuji H, Hirochika H, Fukushi T, Ikeda J. Expression of cauliflower mosaic virus reverse transcriptase in yeast. Nature. 1986;319:240–243. doi: 10.1038/319240a0. - DOI
-
- Rothnie HM, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. - PubMed
-
- Hohn T, Fütterer J. The proteins and functions of plant pararetroviruses : knowns and unknowns. Critical Review in Plant Sciences. 1997;16:133–161.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

