Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development
- PMID: 15831456
- PMCID: PMC1084298
- DOI: 10.1128/MCB.25.9.3492-3505.2005
Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development
Abstract
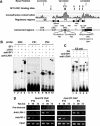
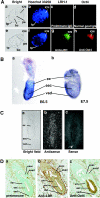
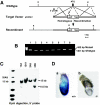
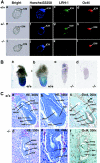
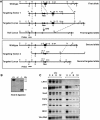
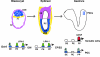
Oct4 plays an essential role in maintaining the inner cell mass and pluripotence of embryonic stem (ES) cells. The expression of Oct4 is regulated by the proximal enhancer and promoter in the epiblast and by the distal enhancer and promoter at all other stages in the pluripotent cell lineage. Here we report that the orphan nuclear receptor LRH-1, which is expressed in undifferentiated ES cells, can bind to SF-1 response elements in the proximal promoter and proximal enhancer of the Oct4 gene and activate Oct4 reporter gene expression. LRH-1 is colocalized with Oct4 in the inner cell mass and the epiblast of embryos at early developmental stages. Disruption of the LRH-1 gene results in loss of Oct4 expression at the epiblast stage and early embryonic death. Using LRH-1(-/-) ES cells, we also show that LRH-1 is required to maintain Oct4 expression at early differentiation time points. In vitro and in vivo results show that LRH-1 plays an essential role in the maintenance of Oct4 expression in ES cells at the epiblast stage of embryonic development, thereby maintaining pluripotence at this crucial developmental stage prior to segregation of the primordial germ cell lineage at gastrulation.
Figures


References
-
- Barnea, E., and Y. Bergman. 2000. Synergy of SF1 and RAR in activation of Oct-3/4 promoter. J. Biol. Chem. 275:6608-6619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
