The distal sequence element of the selenocysteine tRNA gene is a tissue-dependent enhancer essential for mouse embryogenesis
- PMID: 15831471
- PMCID: PMC1084291
- DOI: 10.1128/MCB.25.9.3658-3669.2005
The distal sequence element of the selenocysteine tRNA gene is a tissue-dependent enhancer essential for mouse embryogenesis
Abstract
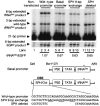
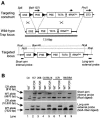
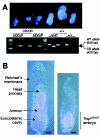
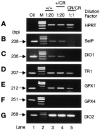
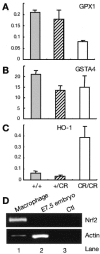
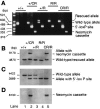
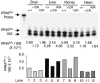
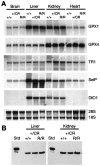
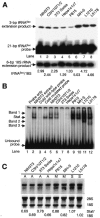
Appropriate expression of the selenocysteine tRNA (tRNA(Sec)) gene is necessary for the production of an entire family of selenoprotein enzymes. This study investigates the consequence of disrupting an upstream enhancer region of the mouse tRNA(Sec) gene (Trsp) known as the distal sequence element (DSE) by use of a conditional repair gene targeting strategy, in which a 3.2-kb insertion was introduced into the promoter of the gene. In the absence of DSE activity, homozygous mice failed to develop in utero beyond embryonic day 7.5 and had severely decreased levels of selenoprotein transcript. Cre-mediated removal of the selection cassette recovered DSE regulation of Trsp, restoring wild-type levels of tRNA(Sec) expression and allowing the generation of viable rescued mice. Further analysis of targeted heterozygous adult mice revealed that the enhancer activity of the DSE is tissue dependent since, in contrast to liver, heart does not require the DSE for normal expression of Trsp. Similarly, in mouse cell lines we showed that the DSE functions as a cell-line-specific inducible element of tRNA(Sec). Together, our data demonstrate that the DSE is a tissue-dependent regulatory element of tRNA(Sec) expression and that its activity is vital for sufficient tRNA(Sec) production during mouse embryogenesis.
Figures
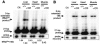
Similar articles
-
The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice.Biochem J. 2009 Feb 15;418(1):61-71. doi: 10.1042/BJ20081304. Biochem J. 2009. PMID: 18973473 Free PMC article.
-
Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development.Molecules. 2009 Sep 10;14(9):3509-27. doi: 10.3390/molecules14093509. Molecules. 2009. PMID: 19783940 Free PMC article. Review.
-
Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium.Mol Cell Biol. 2003 Mar;23(5):1477-88. doi: 10.1128/MCB.23.5.1477-1488.2003. Mol Cell Biol. 2003. PMID: 12588969 Free PMC article.
-
Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34.J Biol Chem. 2007 Nov 9;282(45):32591-602. doi: 10.1074/jbc.M707036200. Epub 2007 Sep 11. J Biol Chem. 2007. PMID: 17848557
-
Selenocysteine tRNAs as central components of selenoprotein biosynthesis in eukaryotes.Biomed Environ Sci. 1997 Sep;10(2-3):116-24. Biomed Environ Sci. 1997. PMID: 9315302 Review.
Cited by
-
Selenoproteins and cardiovascular stress.Thromb Haemost. 2015 Mar;113(3):494-504. doi: 10.1160/TH14-07-0603. Epub 2014 Oct 30. Thromb Haemost. 2015. PMID: 25354851 Free PMC article. Review.
-
The choreography of chromatin in RNA polymerase III regulation.Biochem Soc Trans. 2024 Jun 26;52(3):1173-1189. doi: 10.1042/BST20230770. Biochem Soc Trans. 2024. PMID: 38666598 Free PMC article. Review.
-
The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice.Biochem J. 2009 Feb 15;418(1):61-71. doi: 10.1042/BJ20081304. Biochem J. 2009. PMID: 18973473 Free PMC article.
-
Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation.J Biol Chem. 2011 Jun 3;286(22):19354-63. doi: 10.1074/jbc.M111.219576. Epub 2011 Apr 12. J Biol Chem. 2011. PMID: 21487017 Free PMC article.
-
Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development.Molecules. 2009 Sep 10;14(9):3509-27. doi: 10.3390/molecules14093509. Molecules. 2009. PMID: 19783940 Free PMC article. Review.
References
-
- Adachi, K., H. Saito, T. Tanaka, and T. Oka. 1998. Molecular cloning and characterization of the murine staf cDNA encoding a transcription activating factor for the selenocysteine tRNA gene in mouse mammary gland. J. Biol. Chem. 273:8598-8606. - PubMed
-
- Bianco, A. C., D. Salvatore, B. Gereben, M. J. Berry, and P. R. Larsen. 2002. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocrinol. Rev. 23:38-89. - PubMed
-
- Bosl, M. R., M. F. Seldin, S. Nishimura, and M. Taketo. 1995. Cloning, structural analysis and mapping of the mouse selenocysteine tRNA([Ser]Sec) gene (Trsp). Mol. Gen. Genet. 248:247-252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
