Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog
- PMID: 15831699
- PMCID: PMC1895152
- DOI: 10.1182/blood-2005-02-0707
Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog
Abstract
Jak (Janus kinase) family nonreceptor tyrosine kinases are central mediators of cytokine signaling. The Jak kinases exhibit distinct cytokine receptor association profiles and so transduce different signals. Jak3 expression is limited to the immune system, where it plays a key role in signal transduction from cytokine receptors containing the common gamma-chain, gammac. Patients unable to signal via gammac present with severe combined immunodeficiency (SCID). The finding that Jak3 mutations result in SCID has made it a target for development of lymphocyte-specific immunosuppressants. Here, we present the crystal structure of the Jak3 kinase domain in complex with staurosporine analog AFN941. The kinase domain is in the active conformation, with both activation loop tyrosine residues phosphorylated. The phosphate group on pTyr981 in the activation loop is in part coordinated by an arginine residue in the regulatory C-helix, suggesting a direct mechanism by which the active position of the C-helix is induced by phosphorylation of the activation loop. Such a direct coupling has not been previously observed in tyrosine kinases and may be unique to Jak kinases. The crystal structure provides a detailed view of the Jak3 active site and will facilitate computational and structure-directed approaches to development of Jak3-specific inhibitors.
Figures

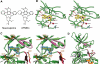
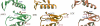
References
-
- Rane SG, Reddy EP. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19: 5662-5679. - PubMed
-
- Imada K, Leonard WJ. The Jak-STAT pathway. Mol Immunol. 2000;37: 1-11. - PubMed
-
- O'Shea JJ, Husa M, Li D, et al. Jak3 and the pathogenesis of severe combined immunodeficiency. Mol Immunol. 2004;41: 727-737. - PubMed
-
- Buckley RH, Schiff RI, Schiff SE, et al. Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130: 378-387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

