Asymmetry in RNA pseudoknots: observation and theory
- PMID: 15831794
- PMCID: PMC1079967
- DOI: 10.1093/nar/gki508
Asymmetry in RNA pseudoknots: observation and theory
Abstract
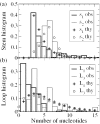
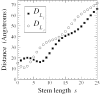
RNA can fold into a topological structure called a pseudoknot, composed of non-nested double-stranded stems connected by single-stranded loops. Our examination of the PseudoBase database of pseudoknotted RNA structures reveals asymmetries in the stem and loop lengths and provocative composition differences between the loops. By taking into account differences between major and minor grooves of the RNA double helix, we explain much of the asymmetry with a simple polymer physics model and statistical mechanical theory, with only one adjustable parameter.
Figures

References
-
- Hofacker I.L., Fontana W., Standler P.F., Bonhoeffer L.S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188.
-
- Lyngsø R.B., Pedersen C.N. RNA pseudoknot prediction in energy-based models. J. Comput. Biol. 2000;7:409–427. - PubMed
-
- Rivas E., Eddy S.R. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

