The microbial selenoproteome of the Sargasso Sea
- PMID: 15833124
- PMCID: PMC1088965
- DOI: 10.1186/gb-2005-6-4-r37
The microbial selenoproteome of the Sargasso Sea
Abstract
Background: Selenocysteine (Sec) is a rare amino acid which occurs in proteins in major domains of life. It is encoded by TGA, which also serves as the signal for termination of translation, precluding identification of selenoprotein genes by available annotation tools. Information on full sets of selenoproteins (selenoproteomes) is essential for understanding the biology of selenium. Herein, we characterized the selenoproteome of the largest microbial sequence dataset, the Sargasso Sea environmental genome project.
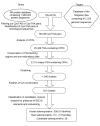
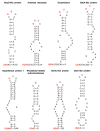
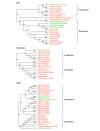
Results: We identified 310 selenoprotein genes that clustered into 25 families, including 101 new selenoprotein genes that belonged to 15 families. Most of these proteins were predicted redox proteins containing catalytic selenocysteines. Several bacterial selenoproteins previously thought to be restricted to eukaryotes were detected by analyzing eukaryotic and bacterial SECIS elements, suggesting that eukaryotic and bacterial selenoprotein sets partially overlapped. The Sargasso Sea microbial selenoproteome was rich in selenoproteins and its composition was different from that observed in the combined set of completely sequenced genomes, suggesting that these genomes do not accurately represent the microbial selenoproteome. Most detected selenoproteins occurred sporadically compared to the widespread presence of their cysteine homologs, suggesting that many selenoproteins recently evolved from cysteine-containing homologs.
Conclusions: This study yielded the largest selenoprotein dataset to date, doubled the number of prokaryotic selenoprotein families and provided insights into forces that drive selenocysteine evolution.
Figures




Similar articles
-
Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues.Genome Biol. 2006;7(10):R94. doi: 10.1186/gb-2006-7-10-r94. Epub 2006 Oct 20. Genome Biol. 2006. PMID: 17054778 Free PMC article.
-
Trends in selenium utilization in marine microbial world revealed through the analysis of the global ocean sampling (GOS) project.PLoS Genet. 2008 Jun 13;4(6):e1000095. doi: 10.1371/journal.pgen.1000095. PLoS Genet. 2008. PMID: 18551170 Free PMC article.
-
The prokaryotic selenoproteome.EMBO Rep. 2004 May;5(5):538-43. doi: 10.1038/sj.embor.7400126. Epub 2004 Apr 23. EMBO Rep. 2004. PMID: 15105824 Free PMC article.
-
Chemical Biology Approaches to Interrogate the Selenoproteome.Acc Chem Res. 2019 Oct 15;52(10):2832-2840. doi: 10.1021/acs.accounts.9b00379. Epub 2019 Sep 16. Acc Chem Res. 2019. PMID: 31523956 Free PMC article. Review.
-
Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine?Exp Cell Res. 2010 May 1;316(8):1296-303. doi: 10.1016/j.yexcr.2010.02.032. Epub 2010 Mar 3. Exp Cell Res. 2010. PMID: 20206159 Review.
Cited by
-
An Ecological Basis for Dual Genetic Code Expansion in Marine Deltaproteobacteria.Front Microbiol. 2021 Jul 14;12:680620. doi: 10.3389/fmicb.2021.680620. eCollection 2021. Front Microbiol. 2021. PMID: 34335502 Free PMC article.
-
Evolution of selenoproteins in the metazoan.BMC Genomics. 2012 Sep 3;13:446. doi: 10.1186/1471-2164-13-446. BMC Genomics. 2012. PMID: 22943432 Free PMC article.
-
A highly efficient form of the selenocysteine insertion sequence element in protozoan parasites and its use in mammalian cells.Proc Natl Acad Sci U S A. 2007 May 8;104(19):7857-62. doi: 10.1073/pnas.0610683104. Epub 2007 Apr 30. Proc Natl Acad Sci U S A. 2007. PMID: 17470795 Free PMC article.
-
High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis.Nucleic Acids Res. 2007;35(15):4952-63. doi: 10.1093/nar/gkm514. Epub 2007 Jul 11. Nucleic Acids Res. 2007. PMID: 17626042 Free PMC article.
-
A method for identification of selenoprotein genes in archaeal genomes.Genomics Proteomics Bioinformatics. 2009 Jun;7(1-2):62-70. doi: 10.1016/S1672-0229(08)60034-0. Genomics Proteomics Bioinformatics. 2009. PMID: 19591793 Free PMC article.
References
-
- Hatfield DL. Selenium Its Molecular Biology and Role in Human Health. Boston: Kluwer Academic Publishers; 2001.
-
- Nirenberg M, Caskey T, Marshall R, Brimacombe R, Kellog D, Doctor BP, Hatfield D, Levin J, Rothman F, Pestka S, et al. The RNA code in protein synthesis. Cold Spring Harbor Symp Quant Biol. 1966;31:11–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

