Derivation of genetic interaction networks from quantitative phenotype data
- PMID: 15833125
- PMCID: PMC1088966
- DOI: 10.1186/gb-2005-6-4-r38
Derivation of genetic interaction networks from quantitative phenotype data
Abstract
We have generalized the derivation of genetic-interaction networks from quantitative phenotype data. Familiar and unfamiliar modes of genetic interaction were identified and defined. A network was derived from agar-invasion phenotypes of mutant yeast. Mutations showed specific modes of genetic interaction with specific biological processes. Mutations formed cliques of significant mutual information in their large-scale patterns of genetic interaction. These local and global interaction patterns reflect the effects of gene perturbations on biological processes and pathways.
Figures
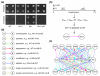
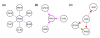

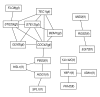
References
-
- Tewari M, Hu PJ, Ahn JS, Ayivi-Guedehoussou N, Vidalain PO, Li S, Milstein S, Armstrong CM, Boxem M, Butler MD, et al. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-beta signaling network. Mol Cell. 2004;13:469–482. doi: 10.1016/S1097-2765(04)00033-4. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

