Filtering high-throughput protein-protein interaction data using a combination of genomic features
- PMID: 15833142
- PMCID: PMC1127019
- DOI: 10.1186/1471-2105-6-100
Filtering high-throughput protein-protein interaction data using a combination of genomic features
Abstract
Background: Protein-protein interaction data used in the creation or prediction of molecular networks is usually obtained from large scale or high-throughput experiments. This experimental data is liable to contain a large number of spurious interactions. Hence, there is a need to validate the interactions and filter out the incorrect data before using them in prediction studies.
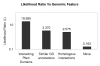
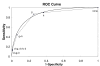
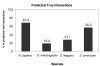
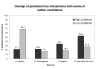
Results: In this study, we use a combination of 3 genomic features -- structurally known interacting Pfam domains, Gene Ontology annotations and sequence homology -- as a means to assign reliability to the protein-protein interactions in Saccharomyces cerevisiae determined by high-throughput experiments. Using Bayesian network approaches, we show that protein-protein interactions from high-throughput data supported by one or more genomic features have a higher likelihood ratio and hence are more likely to be real interactions. Our method has a high sensitivity (90%) and good specificity (63%). We show that 56% of the interactions from high-throughput experiments in Saccharomyces cerevisiae have high reliability. We use the method to estimate the number of true interactions in the high-throughput protein-protein interaction data sets in Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens to be 27%, 18% and 68% respectively. Our results are available for searching and downloading at http://helix.protein.osaka-u.ac.jp/htp/.
Conclusion: A combination of genomic features that include sequence, structure and annotation information is a good predictor of true interactions in large and noisy high-throughput data sets. The method has a very high sensitivity and good specificity and can be used to assign a likelihood ratio, corresponding to the reliability, to each interaction.
Figures
References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
-
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

