Toward a neutral evolutionary model of gene expression
- PMID: 15834146
- PMCID: PMC1450413
- DOI: 10.1534/genetics.104.037135
Toward a neutral evolutionary model of gene expression
Abstract
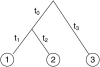
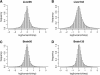
We introduce a stochastic model that describes neutral changes of gene expression over evolutionary time as a compound Poisson process where evolutionary events cause changes of expression level according to a given probability distribution. The model produces simple estimators for model parameters and allows discrimination between symmetric and asymmetric distributions of evolutionary expression changes along an evolutionary lineage. Furthermore, we introduce two measures, the skewness of expression difference distributions and relative difference of evolutionary branch lengths, which are used to quantify deviation from clock-like behavior of gene expression distances. Model-based analyses of gene expression profiles in primate liver and brain samples yield the following results: (1) The majority of gene expression changes are consistent with a neutral model of evolution; (2) along evolutionary lineages, upward changes in expression are less frequent but of greater average magnitude than downward changes; and (3) the skewness measure and the relative branch length difference confirm that an acceleration of gene expression evolution occurred on the human lineage in brain but not in liver. We discuss the latter result with respect to a neutral model of transcriptome evolution and show that a small number of genes expressed in brain can account for the observed data.
Figures
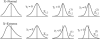


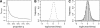
Similar articles
-
A neutral model of transcriptome evolution.PLoS Biol. 2004 May;2(5):E132. doi: 10.1371/journal.pbio.0020132. Epub 2004 May 11. PLoS Biol. 2004. PMID: 15138501 Free PMC article.
-
Phylogenetic reconstruction of ancestral character states for gene expression and mRNA splicing data.BMC Bioinformatics. 2005 May 27;6:127. doi: 10.1186/1471-2105-6-127. BMC Bioinformatics. 2005. PMID: 15921519 Free PMC article.
-
Hominoid phylogeny estimated by model selection using goodness of fit significance tests.Mol Phylogenet Evol. 1995 Sep;4(3):283-90. doi: 10.1006/mpev.1995.1025. Mol Phylogenet Evol. 1995. PMID: 8845964
-
Evolution of primate gene expression.Nat Rev Genet. 2006 Sep;7(9):693-702. doi: 10.1038/nrg1940. Nat Rev Genet. 2006. PMID: 16921347 Review.
-
The molecular clock and evolutionary timescales.Biochem Soc Trans. 2018 Oct 19;46(5):1183-1190. doi: 10.1042/BST20180186. Epub 2018 Aug 28. Biochem Soc Trans. 2018. PMID: 30154097 Review.
Cited by
-
The new mutation theory of phenotypic evolution.Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12235-42. doi: 10.1073/pnas.0703349104. Epub 2007 Jul 17. Proc Natl Acad Sci U S A. 2007. PMID: 17640887 Free PMC article.
-
On the specificity of gene regulatory networks: How does network co-option affect subsequent evolution?Curr Top Dev Biol. 2020;139:375-405. doi: 10.1016/bs.ctdb.2020.03.002. Epub 2020 May 5. Curr Top Dev Biol. 2020. PMID: 32450967 Free PMC article. Review.
-
Gene Expression Variation Resolves Species and Individual Strains among Coral-Associated Dinoflagellates within the Genus Symbiodinium.Genome Biol Evol. 2016 Feb 11;8(3):665-80. doi: 10.1093/gbe/evw019. Genome Biol Evol. 2016. PMID: 26868597 Free PMC article.
-
Sensitivity of quantitative traits to mutational effects and number of loci.Theor Popul Biol. 2015 Jun;102:85-93. doi: 10.1016/j.tpb.2015.03.005. Epub 2015 Mar 31. Theor Popul Biol. 2015. PMID: 25840144 Free PMC article.
-
Efficient Recycled Algorithms for Quantitative Trait Models on Phylogenies.Genome Biol Evol. 2016 May 12;8(5):1338-50. doi: 10.1093/gbe/evw064. Genome Biol Evol. 2016. PMID: 27056412 Free PMC article.
References
-
- Balding, D. J., M. Bishop and C. Cannings, 2003 Handbook of Statistical Genetics. John Wiley & Sons, Chichester, UK.
-
- Bolstad, B. M., R. A. Irizarry, M. Astrand and T. P. Speed, 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. - PubMed
-
- Cheung, V. G., L. K. Conlin, T. M. Weber, M. Arcaro, K. Y. Jen et al., 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 33: 422–425. - PubMed
-
- Durrett, R., 2002 Probability Models for DNA Sequence Evolution. Springer, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

