A developmental switch in the response of DRG neurons to ETS transcription factor signaling
- PMID: 15836427
- PMCID: PMC1084331
- DOI: 10.1371/journal.pbio.0030159
A developmental switch in the response of DRG neurons to ETS transcription factor signaling
Abstract
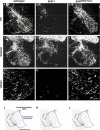
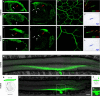
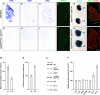
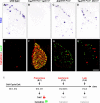
Two ETS transcription factors of the Pea3 subfamily are induced in subpopulations of dorsal root ganglion (DRG) sensory and spinal motor neurons by target-derived factors. Their expression controls late aspects of neuronal differentiation such as target invasion and branching. Here, we show that the late onset of ETS gene expression is an essential requirement for normal sensory neuron differentiation. We provide genetic evidence in the mouse that precocious ETS expression in DRG sensory neurons perturbs axonal projections, the acquisition of terminal differentiation markers, and their dependence on neurotrophic support. Together, our findings indicate that DRG sensory neurons exhibit a temporal developmental switch that can be revealed by distinct responses to ETS transcription factor signaling at sequential steps of neuronal maturation.
Figures




References
-
- Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: A view from the nervous system. Cell. 1999;96:211–224. - PubMed
-
- Anderson DJ, Groves A, Lo L, Ma Q, Rao M, et al. Cell lineage determination and the control of neuronal identity in the neural crest. Cold Spring Harb Symp Quant Biol. 1997;62:493–504. - PubMed
-
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: A multigene process. Nat Rev Genet. 2002;3:453–461. - PubMed
-
- Bibel M, Barde YA. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

