Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus
- PMID: 15836513
- PMCID: PMC3523328
- DOI: 10.1111/j.1399-302X.2004.00190.x
Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus
Abstract
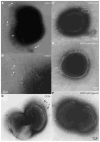
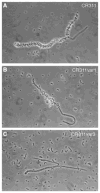
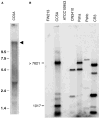
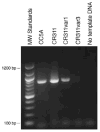
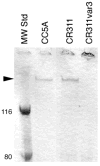
The surface of the oral plaque bacterium Streptococcus cristatus is decorated with a lateral tuft of fibrils. The fibrillar tuft functions in the adhesion of S. cristatus to heterologous bacterial species in the plaque biofilm. The tuft typically consists of a densely packed fringe of shorter fibrils 238 +/- 19 nm long with longer, less abundant fibrils 403 +/- 66 nm long projecting through the fringe of short fibrils. The two types of fibrils in the tufts of S. cristatus have been refractory to biochemical separation, complicating their characterization. A hexadecane partition assay was used to enrich for subpopulations of S. cristatus CR311 (type strain NCTC 12479) having distinct fibrillar morphotypes. Negative staining in the TEM revealed that cells of a hydrophobic subpopulation of S. cristatus (CR311var1) carried only the long fibrils (395 +/- 32 nm). A hydrophilic subpopulation of S. cristatus (CR311var3) consisted of mixed morphotypes having no fibrils or remnant short fibrils (223 +/- 49 nm). No long fibrils were observed on any cells in the CR311var3 subpopulation. The CR311var3 morphotype, unlike the wild-type strain and CR311var1, was not able to form corncobs with either Corynebacterium matruchotii or Fusobacterium nucleatum. Variant CR311var3 did not express the novel gene srpA, which encodes a high molecular weight (321,882 Da) serine-rich protein, SrpA. The SrpA protein contains two extensive repeat motifs of 17 and 71 amino acids and a gram-positive cell wall anchor consensus sequence (LPNTG). The unusual properties of SrpA most closely resemble those of Fap1, the fimbrial-associated adhesin protein of Streptococcus parasanguis. The association of long fibrils, high surface hydrophobicity, ability to form corncob formations, and expression of the srpA gene suggest that SrpA is a long fibril protein in S. cristatus.
Figures


References
-
- Busscher HJ, Handley PS, Rouxhet P, Hesketh GLM, van der Mei HC. The relationship between structural and physicochemical surface properties of tufted Streptococcus sanguis strains. In: Mozes N, Busscher HJ, Handley PS, Busscher HJ, Rouxhet PG, editors. Microbial Cell Surface Analysis, Structural and Physicochemical Methods. Weinheim; Wiley-VCH: 1991. pp. 319–337.
-
- Correia FF, Lamont RJ, Bayer ME, Rosan B, DiRienzo JM. Cloning and sequencing of a mutated locus that affects fimbrial tuft organization and corncob formation in Streptococcus crista CC5A. Int J Oral Biol. 1997;22:241–248.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

