Cis-acting expression quantitative trait loci in mice
- PMID: 15837804
- PMCID: PMC1088296
- DOI: 10.1101/gr.3216905
Cis-acting expression quantitative trait loci in mice
Abstract
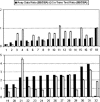
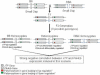
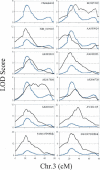
We previously reported the analysis of genome-wide expression profiles and various diabetes-related traits in a segregating cross between inbred mouse strains C57BL/6J (B6) and DBA/2J (DBA). By considering transcript levels as quantitative traits, we identified several thousand expression quantitative trait loci (eQTL) with LOD score >4.3. We now experimentally address the problem of multiple comparisons by estimating the fraction of false-positive eQTL that are under cis-acting regulation. For this, we have utilized a classic cis-trans test with (B6 x DBA)F(1) mice to determine the relative levels of transcripts from the B6 and DBA alleles. The results suggest that at least 64% of cis-acting eQTL with LOD >4.3 are true positives, while the remaining 36% could not be confirmed as truly cis-acting. Moreover, we find that >96% of apparent cis-acting eQTL occur in regions that do not share SNP haplotypes. Cis-acting eQTL serve as an important new resource for the identification of positional candidates in QTL studies in mice. Also, we use the analysis of the correlation structures between genotypes, gene expression traits, and phenotypic traits to further characterize genes expressed in liver that are under cis-acting control, and highlight the advantages and disadvantages of integrating genetics and gene expression data in segregating populations.
Figures



References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. J. Mol. Biol. 215: 403–410. - PubMed
-
- Brem, R.B., Yvert, G., Clinton, R., and Kruglyak, L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. - PubMed
-
- Chesler, E.J., Wang, J., Lu, L., Qu, Y., Manly, K.F., and Williams, R.W. 2003. Genetic correlates of gene expression in recombinant inbred strains: A relational model system to explore neurobehavioral phenotypes. Neuroinformatics 1: 343–357. - PubMed
-
- Colinayo, V.V., Qiao, J.H., Wang, X., Krass, K.L., Schadt, E., Lusis, A.J., and Drake, T.A. 2003. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mammalian Genome 14: 464–471. - PubMed
Web site references
-
- http://www.pyrosequencing.com; Pyrosequencing SNP primer design software v1.0.1.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
