The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation
- PMID: 15837807
- PMCID: PMC1088291
- DOI: 10.1101/gr.3684805
The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation
Abstract
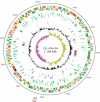
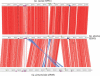
The obligate intracellular bacterial pathogen Chlamydophila abortus strain S26/3 (formerly the abortion subtype of Chlamydia psittaci) is an important cause of late gestation abortions in ruminants and pigs. Furthermore, although relatively rare, zoonotic infection can result in acute illness and miscarriage in pregnant women. The complete genome sequence was determined and shows a high level of conservation in both sequence and overall gene content in comparison to other Chlamydiaceae. The 1,144,377-bp genome contains 961 predicted coding sequences, 842 of which are conserved with those of Chlamydophila caviae and Chlamydophila pneumoniae. Within this conserved Cp. abortus core genome we have identified the major regions of variation and have focused our analysis on these loci, several of which were found to encode highly variable protein families, such as TMH/Inc and Pmp families, which are strong candidates for the source of diversity in host tropism and disease causation in this group of organisms. Significantly, Cp. abortus lacks any toxin genes, and also lacks genes involved in tryptophan metabolism and nucleotide salvaging (guaB is present as a pseudogene), suggesting that the genetic basis of niche adaptation of this species is distinct from those previously proposed for other chlamydial species.
Figures



Similar articles
-
Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae.Nucleic Acids Res. 2003 Apr 15;31(8):2134-47. doi: 10.1093/nar/gkg321. Nucleic Acids Res. 2003. PMID: 12682364 Free PMC article.
-
Chlamydiaceae Genomics Reveals Interspecies Admixture and the Recent Evolution of Chlamydia abortus Infecting Lower Mammalian Species and Humans.Genome Biol Evol. 2015 Oct 27;7(11):3070-84. doi: 10.1093/gbe/evv201. Genome Biol Evol. 2015. PMID: 26507799 Free PMC article.
-
European Chlamydia abortus livestock isolate genomes reveal unusual stability and limited diversity, reflected in geographical signatures.BMC Genomics. 2017 May 4;18(1):344. doi: 10.1186/s12864-017-3657-y. BMC Genomics. 2017. PMID: 28472926 Free PMC article.
-
Genome sequence of the Chlamydophila abortus variant strain LLG.J Bacteriol. 2011 Aug;193(16):4276-7. doi: 10.1128/JB.05290-11. Epub 2011 Jun 17. J Bacteriol. 2011. PMID: 21685275 Free PMC article.
-
[Chlamydial diseases of domestic animals--zoonotic potential of the agents and diagnostic issues].Dtsch Tierarztl Wochenschr. 2002 Apr;109(4):142-8. Dtsch Tierarztl Wochenschr. 2002. PMID: 11998363 Review. German.
Cited by
-
Identification of immunologically relevant proteins of Chlamydophila abortus using sera from experimentally infected pregnant ewes.Clin Vaccine Immunol. 2010 Aug;17(8):1274-81. doi: 10.1128/CVI.00163-10. Epub 2010 Jun 16. Clin Vaccine Immunol. 2010. PMID: 20554807 Free PMC article.
-
Pathogenic Bacillus anthracis in the progressive gene losses and gains in adaptive evolution.BMC Bioinformatics. 2009 Jan 30;10 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2105-10-S1-S3. BMC Bioinformatics. 2009. PMID: 19208130 Free PMC article.
-
Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis.BMC Microbiol. 2008 Feb 28;8:42. doi: 10.1186/1471-2180-8-42. BMC Microbiol. 2008. PMID: 18307777 Free PMC article.
-
Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism.Future Microbiol. 2013 Sep;8(9):1129-1146. doi: 10.2217/fmb.13.80. Future Microbiol. 2013. PMID: 24020741 Free PMC article. Review.
-
Chlamydia Infection Remodels Host Cell Mitochondria to Alter Energy Metabolism and Subvert Apoptosis.Microorganisms. 2023 May 24;11(6):1382. doi: 10.3390/microorganisms11061382. Microorganisms. 2023. PMID: 37374883 Free PMC article. Review.
References
-
- Andersson, S.G.E., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C.M., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140. - PubMed
-
- Bannantine, J.P. and Stabel, J.R. 1999. Identification of Mycobacterium paratuberculosis antigens present within infected macrophages. Mol. Biol. Cell 10: 1047.
-
- Bannantine, J.P., Rockey, D.D., and Hackstadt, T. 1998a. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 28: 1017–1026. - PubMed
Web site references
-
- http://hmmer.wustl.edu/; profile hidden Markov models for biological sequence analysis.
-
- http://www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html; Belvu software for Multiple sequence alignment.
-
- http://www.sanger.ac.uk/Software/ACT/; ACT comparative sequence analysis tool.
-
- http://www.sanger.ac.uk/Software/Artemis/; Artemis sequence annotation tool.
-
- http://www.sanger.ac.uk/Software/Pfam/tsearch.shtml;; PFAM protein family database.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
