Role of HtrA in growth and competence of Streptococcus mutans UA159
- PMID: 15838029
- PMCID: PMC1082816
- DOI: 10.1128/JB.187.9.3028-3038.2005
Role of HtrA in growth and competence of Streptococcus mutans UA159
Abstract
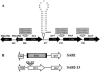
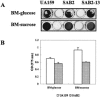
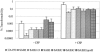
We report here that HtrA plays a role in controlling growth and competence development for genetic transformation in Streptococcus mutans. Disruption of the gene for HtrA resulted in slow growth at 37 degrees C, reduced thermal tolerance at 42 degrees C, and altered sucrose-dependent biofilm formation on polystyrene surfaces. The htrA mutant also displayed a significantly reduced ability to undergo genetic transformation. A direct association between HtrA and genetic competence was demonstrated by the increased expression of the htrA gene upon exposure to competence-stimulating peptide. The induction of htrA gradually reached a maximum at around 20 min, suggesting that HtrA may be involved in a late competence response. Complementation of the htrA mutation in a single copy on the chromosome of the mutant could rescue the defective growth phenotypes but not transformability, apparently because a second gene, spo0J, immediately downstream of htrA, also affects transformation. The htrA and spo0J genes were shown to be both individually transcribed and cotranscribed and probably have a functional connection in competence development. HtrA regulation appears to be finely tuned in S. mutans, since strains containing multiple copies of htrA exhibited abnormal growth phenotypes. Collectively, the results reveal HtrA to be an integral component of the regulatory network connecting cellular growth, stress tolerance, biofilm formation, and competence development and reveal a novel role for the spo0J gene in genetic transformation.
Figures



Similar articles
-
Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo.Infect Immun. 2007 Apr;75(4):1679-89. doi: 10.1128/IAI.01581-06. Epub 2007 Jan 12. Infect Immun. 2007. PMID: 17220310 Free PMC article.
-
Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae.J Bacteriol. 2004 Aug;186(16):5258-66. doi: 10.1128/JB.186.16.5258-5266.2004. J Bacteriol. 2004. PMID: 15292127 Free PMC article.
-
Role of HtrA in surface protein expression and biofilm formation by Streptococcus mutans.Infect Immun. 2005 Oct;73(10):6923-34. doi: 10.1128/IAI.73.10.6923-6934.2005. Infect Immun. 2005. PMID: 16177372 Free PMC article.
-
Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159.Infect Immun. 2006 Mar;74(3):1631-42. doi: 10.1128/IAI.74.3.1631-1642.2006. Infect Immun. 2006. PMID: 16495534 Free PMC article.
-
HtrA serine proteases as potential therapeutic targets in cancer.Curr Cancer Drug Targets. 2009 Jun;9(4):451-68. doi: 10.2174/156800909788486704. Curr Cancer Drug Targets. 2009. PMID: 19519315 Free PMC article. Review.
Cited by
-
The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis.J Bacteriol. 2009 Apr;191(7):2248-56. doi: 10.1128/JB.01726-08. Epub 2009 Jan 23. J Bacteriol. 2009. PMID: 19168608 Free PMC article.
-
ciaR impacts biofilm formation by regulating an arginine biosynthesis pathway in Streptococcus sanguinis SK36.Sci Rep. 2017 Dec 7;7(1):17183. doi: 10.1038/s41598-017-17383-1. Sci Rep. 2017. PMID: 29215019 Free PMC article.
-
clpB, a class III heat-shock gene regulated by CtsR, is involved in thermotolerance and virulence of Enterococcus faecalis.Microbiology (Reading). 2011 Mar;157(Pt 3):656-665. doi: 10.1099/mic.0.041897-0. Epub 2010 Dec 9. Microbiology (Reading). 2011. PMID: 21148206 Free PMC article.
-
A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans.J Bacteriol. 2011 Feb;193(4):862-74. doi: 10.1128/JB.01257-10. Epub 2010 Dec 10. J Bacteriol. 2011. PMID: 21148727 Free PMC article.
-
Mechanistic, genomic and proteomic study on the effects of BisGMA-derived biodegradation product on cariogenic bacteria.Dent Mater. 2017 Feb;33(2):175-190. doi: 10.1016/j.dental.2016.11.007. Epub 2016 Dec 3. Dent Mater. 2017. PMID: 27919444 Free PMC article.
References
-
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. - PMC - PubMed
-
- Allaker, R. P., S. V. Seddon, C. Tredwin, and E. Lynch. 1998. Detection of Streptococcus mutans by PCR amplification of the spaP gene in teeth rendered caries free. J. Dent. 26:443-445. - PubMed
-
- Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

